Autophagic Flux Assay Kit

Autophagic Flux Assay Kit (Autophagy)
- Easy to analyze the autophagy pathway just by adding the reagent
- All-in-one Kit with Lysosome Acidification Inhibitor
-
Product codeA562 Autophagic Flux Assay Kit
| Unit size | Price | Item Code |
|---|---|---|
| 1set | Find your distributors | A562-10 |
| 1set | DALGreen DAPRed Bafilomycin A1 |
x 1 x 1 x 1 |
|---|
Description
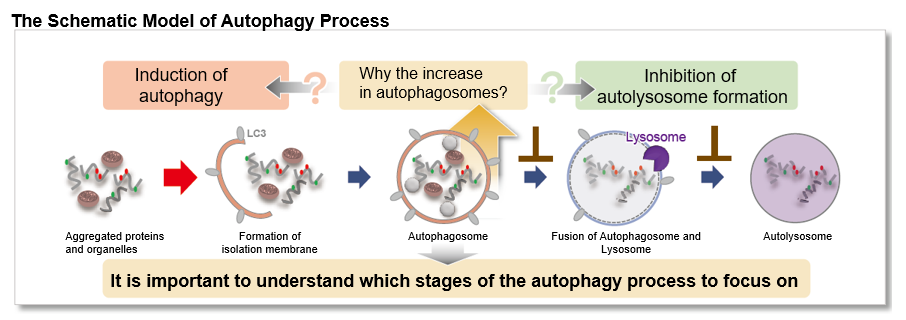
Autophagy, the regulated degradation process of cytoplasmic components such as proteins and organelles has been actively studied in aging and neurodegenerative diseases. There is a growing need to determine whether the increase in autophagosomes is due to autophagy induction or autolysosome inhibition. Understanding the autophagic flux is crucial.
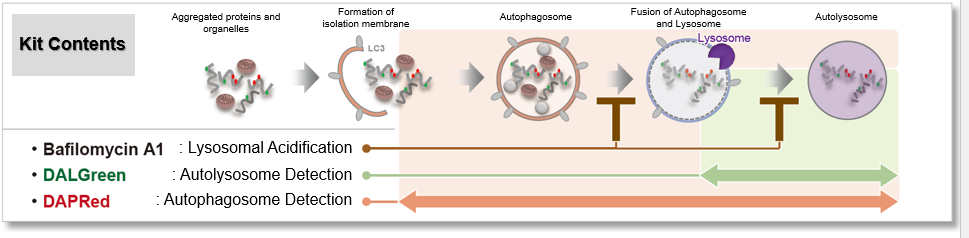
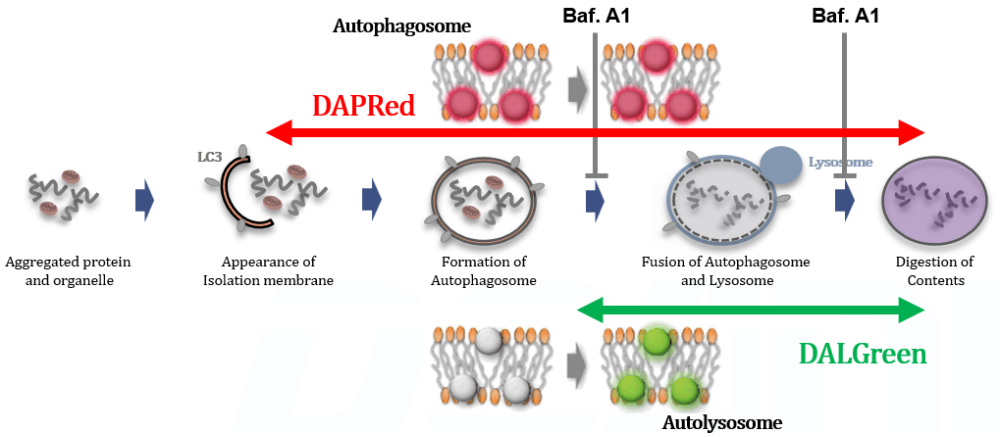
This kit contains autophagosome and autolysosome detection dye (DAPRed), autolysosome detection dye (DALGreen), and lysosomal acidification inhibitor (bafilomycin A1). The Autophagic Flux Assay Kit allows the accurate evaluation of autophagic flux by monitoring autophagosome formation, lysosome fusion, and digestion of contents1).
1) H. Sakurai, et al., "Development of small fluorescent probes for the analysis of autophagy kinetics“, iScience, 2023, 26, 107218.
Autophagy Analysis Products
| Product Name | Autophagosome Detection Dyes and Fluorescence Properties |
Autolysosome Detection Dyes and Fluorescence Properties |
|---|---|---|
| Autophagic Flux Assay Kit* | DAPRed Ex: 500-560 nm / Em: 690-750 nm |
DALGreen Ex: 350-450 nm / Em: 500-560 nm |
| DAPGreen - Autophagy Detection | DAPGreen Ex: 425-475 nm / Em: 500-560 nm |
|
| DAPRed - Autophagy Detection | DAPRed Ex: 500-560 nm / Em: 690-750 nm |
|
| DALGreen - Autophagy Detection | DALGreen Ex: 350-450 nm / Em: 500-560 nm |
*Kit includes Lysosome Acidification Inhibitor
Manual
Technical info
This kit contains DAPRed (DAPRed - Autophagy Detection) for the detection of autophagosomes and autolysosomes, DALGreen (DALGreen - Autophagy Detection) for the detection of autolysosomes, and Bafilomycin A1 for inhibiting lysosome acidification. By simply adding the reagents, you can monitor the process from autophagosome formation to autolysosome formation 2,3).
2) X. Chen, et al, "Chaiqi decoction ameliorates vascular endothelial injury in metabolic syndrome by upregulating autophagy, Am J Transl Res., 2020, 12(9), 4902-4922.
3) C. Oh, et al, "S-Nitrosylation of p62 Inhibits Autophagic Flux to Promote α-Synuclein Secretion and Spread in Parkinson's Disease and Lewy Body Dementia, J Neurosci., 2022, 42(14), 3011-3024.
Experimental example 1
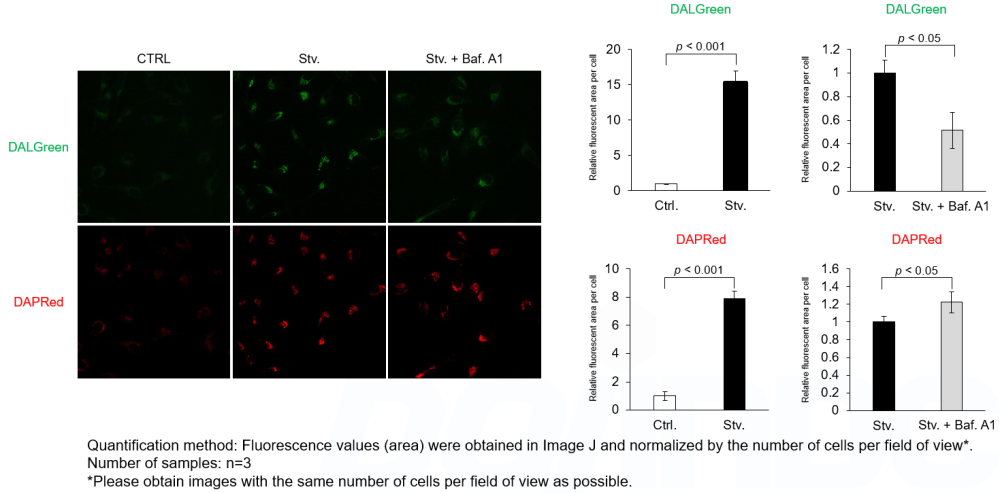
Analysis of autolysosome formation inhibition using Bafilomycin A1 by confocal fluorescence microscopy
DALGreen and DAPRed labeled HeLa cells were used to evaluate changes in autophagic flux induced by the lysosomal acidification inhibitor bafilomycin A1 (Baf. A1). Compared to starvation conditions, the fluorescence signals of DALGreen were decreased under inhibited conditions of autolysosome formation by the addition of Baf. A1. In contrast, the fluorescence signals of DAPRed were increased under the same conditions, indicating that Baf. A1 led to the accumulation of autophagosome.
*Imaging analysis may be affected by the Baf. A1 dilution rate, duration of stimulation, and timing of addition. Please refer to the Q&A on the product web page for Baf. A1 stimulation.
<Experimental Conditions>
CTRL: Normal condition, Stv.: Induction of autophagy, Stv. + Baf. A1: Inhibition of autolysosome formation
DALGreen filter set: 488 nm (Ex), 490–550 nm (Em)
DAPRed filter set: 561 nm (Ex), 565–700 nm (Em)
<Procedure>
1. HeLa cells were seeded (1.0 x 104 cells/well) on a μ-slide 8 well plate (ibidi) and cultured overnight at 37°C in an incubator equilibrated with 95% air and 5% CO2.
2. After washing twice with MEM containing 10% fetal bovine serum, 200 μl of DALGreen/DAPRed working solution (DALGreen: 1 µmol/l, DAPRed: 0.2 µmol/l) and the cells were incubated at 37°C for 30 minutes.
3. The supernatant was discarded, and the cells were washed twice with MEM containing 10% fetal bovine serum.
4. Samples were prepared under the following conditions.
• MEM containing 10% fetal bovine serum (200 µl) was added to the well, and the cells were incubated at 37 °C for 2 hours 20 minutes. (Control)
• Amino acid-free medium (FUJIFILM Wako Pure Chemical Industries, Ltd., Catalogue code: 048-33575) (200 μl) was added to the well, and the cells were incubated at 37°C for 2 hours 20 minutes. (Starvation)
• Amino acid-free medium (200 μl) was added to the well, and the cells were incubated at 37°C for 2 hours. The supernatant was discarded, bafilomycin A1 working solution (2,000 times dilution, 200 μl), an inhibitor of lysosomal acidification, was added to the well, and the cells were incubated at 37°C for 20 minutes. (Inhibition of autolysosome formation)
5. The stained cells were observed under a confocal fluorescence microscope.
Experimental example 2
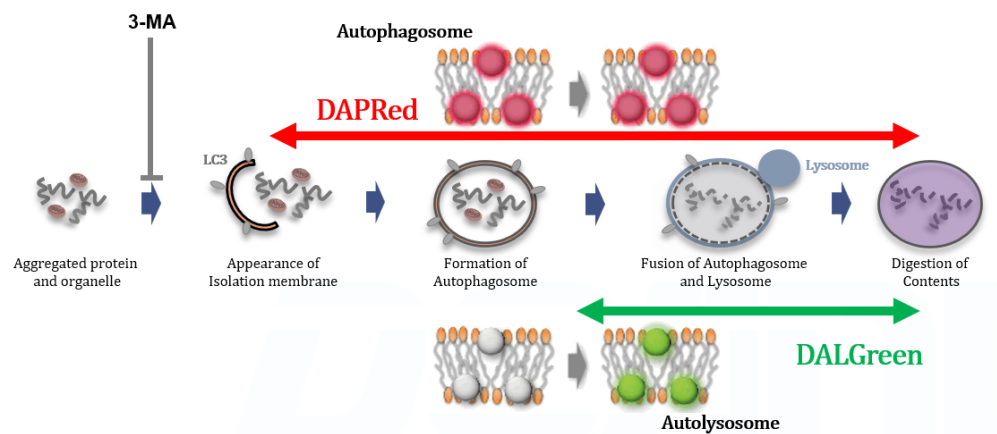
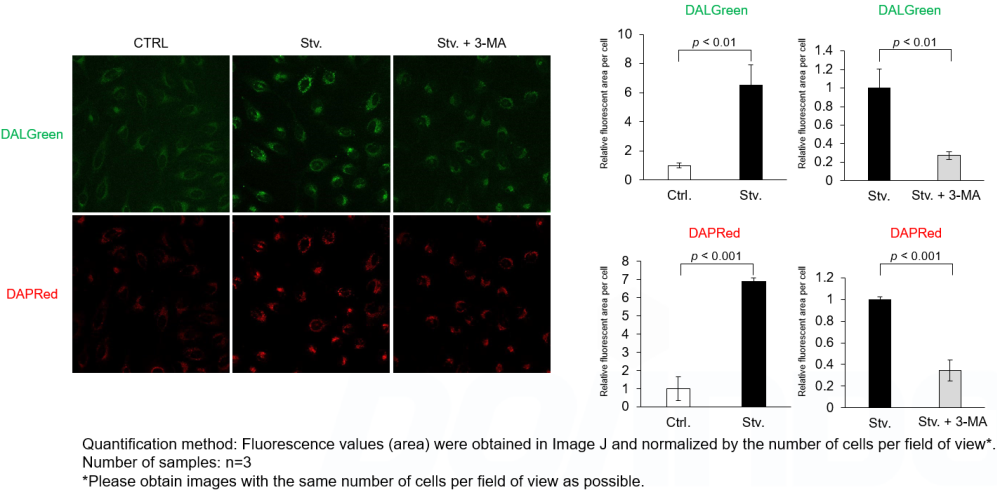
Analysis of autophagosome formation inhibition using 3-MA by confocal fluorescence microscopy
DALGreen and DAPRed labeled HeLa cells were used to evaluate changes in autophagic flux induced by the autophagy inhibitor 3-MA. Compared to starvation conditions, the fluorescence signals of DALGreen and DAPRed were decreased under inhibited conditions of autophagosome formation by the addition of 3-MA.
*The results may be affected by the 3-MA duration of stimulation, and timing of addition. Please refer to the Q&A on the product web page for 3-MA stimulation.
<Experimental Conditions>
CTRL: Normal condition, Stv.: Induction of autophagy, Stv. + 3-MA: Inhibition of autophagosome formation
DALGreen filter set: 488 nm (Ex), 490–550 nm (Em)
DAPRed filter set: 561 nm (Ex), 565–700 nm (Em)
<Procedure>
1. HeLa cells were seeded (1.0 x 104 cells/well) on a μ-slide 8 well plate (ibidi) and cultured overnight at 37°C in an incubator equilibrated with 95% air and 5% CO2.
2. After washing twice with MEM containing 10% fetal bovine serum, 200 μl of DALGreen/DAPRed working solution (DALGreen: 1 µmol/l, DAPRed: 0.2 µmol/l) and the cells were incubated at 37°C for 30 minutes.
3. The supernatant was discarded, and the cells were washed twice with MEM containing 10% fetal bovine serum.
4. Samples were prepared under the following conditions.
• MEM containing 10% fetal bovine serum (200 µl) was added to the well, and the cells were incubated at 37 °C for 2 hours 20 minutes. (Control)
• Amino acid-free medium (FUJIFILM Wako Pure Chemical Industries, Ltd., Catalogue code: 048-33575) (200 μl) was added to the well, and the cells were incubated at 37°C for 2 hours 20 minutes. (Starvation)
• Amino acid-free medium (200 μl) was added to the well, and the cells were incubated at 37°C for 2 hours. The supernatant was discarded, amino acid-free medium containing 3-MA (10 mmol/l, 200 μl), an inhibitor of autophagy, was added to the well, and the cells were incubated at 37°C for 20 minutes. (Inhibition of autophagosome formation)
5. The stained cells were observed under a confocal fluorescence microscope.
Experimental example 3
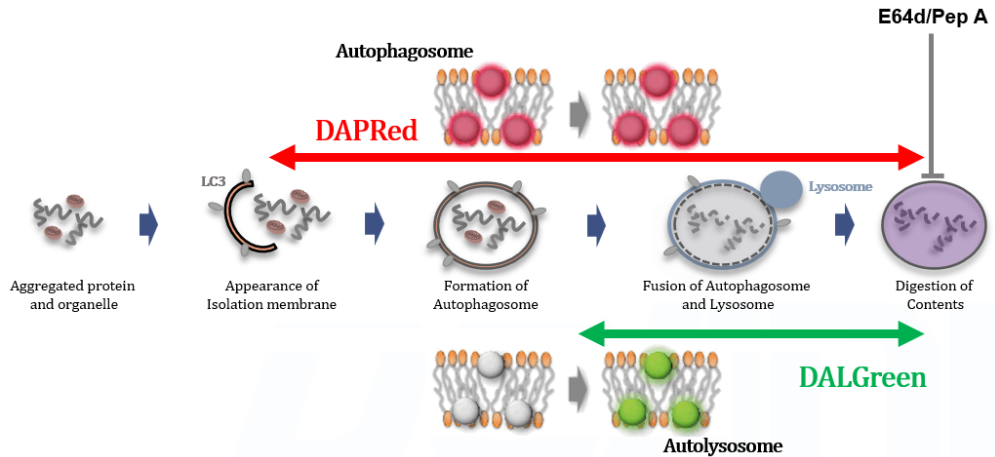
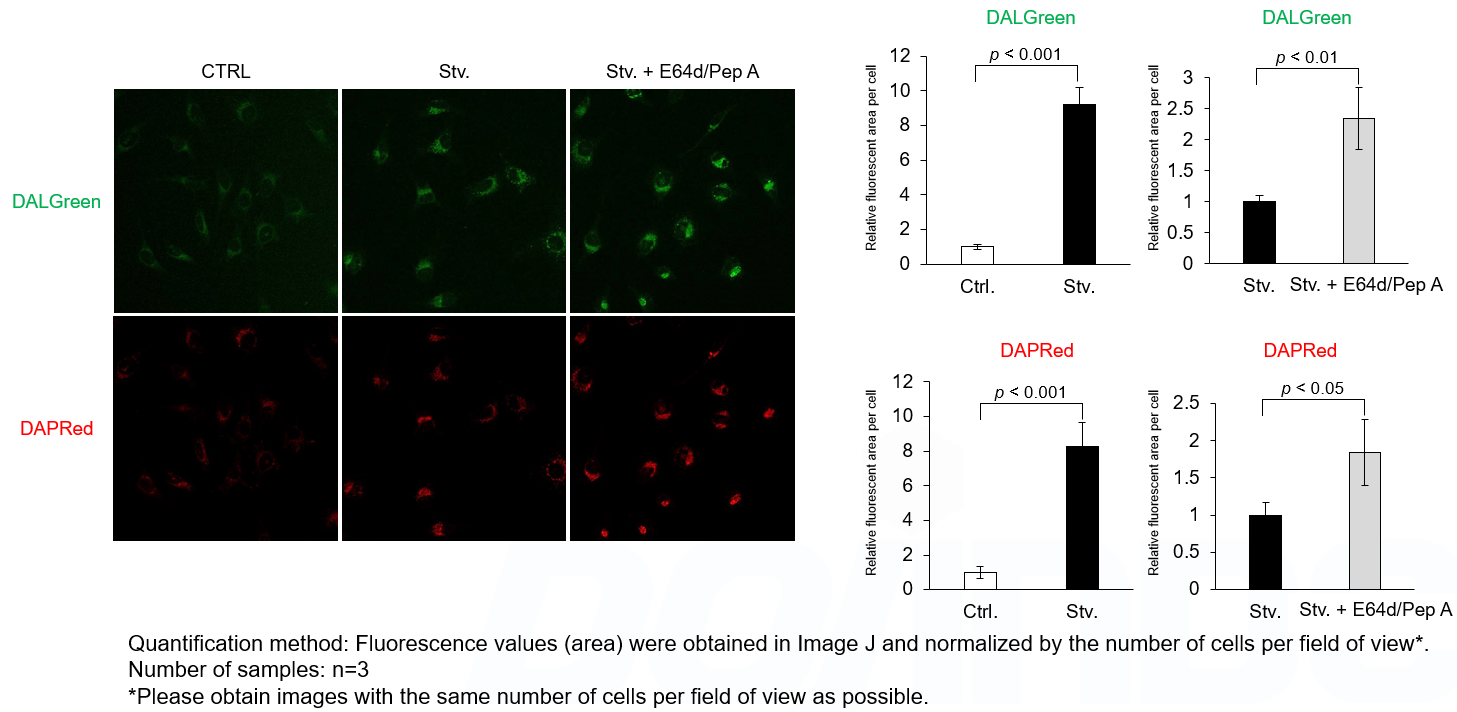
Analysis of lysosomal protein degradation inhibition using E64d/Pepstatin A by confocal fluorescence microscopy
DALGreen and DAPRed labeled HeLa cells were used to evaluate changes in autophagic flux induced by the lysosomal protease inhibitor E64d/Pepstatin A (Pep A). Compared to starvation conditions, the fluorescence signals of DALGreen and DAPRed were increased due to autolysosome accumulation by the addition of E64d/Pep A.
<Experimental Conditions>
CTRL: Normal condition, Stv.: Induction of autophagy, Stv. + E64d/Pep A: Inhibition of lysosomal protein degradation
DALGreen filter set: 488 nm (Ex), 490–550 nm (Em)
DAPRed filter set: 561 nm (Ex), 565–700 nm (Em)
<Procedure>
1. HeLa cells were seeded (1.0 x 104 cells/well) on a μ-slide 8 well plate (ibidi) and cultured overnight at 37 °C in an incubator equilibrated with 95% air and 5% CO2.
2. After washing twice with MEM containing 10% fetal bovine serum, 200 μl of DALGreen/DAPRed working solution (DALGreen: 1 µmol/l, DAPRed: 0.2 µmol/l) and the cells were incubated at 37°C for 30 minutes.
3. The supernatant was discarded, and the cells were washed twice with MEM containing 10% fetal bovine serum.
4. Samples were prepared under the following conditions.
• MEM containing 10% fetal bovine serum (200 µl) was added to the well, and the cells were incubated at 37°C for 2 hours. (Control)
• Amino acid-free medium (FUJIFILM Wako Pure Chemical Industries, Ltd., Catalogue code: 048-33575) (200 μl) was added to the well, and the cells were incubated at 37°C for 2 hours. (Starvation)
• Amino acid-free medium containing E64d/Pep A (10 µg/ml each, 200 μl), an inhibitor of lysosomal protease, was added to the well, and the cells were incubated at 37°C for 2 hours. (Inhibition of lysosomal protein degradation)
5. The stained cells were observed under a confocal fluorescence microscope.
Comparative Table of Autophagy Detection Reagents
We have a lineup of kits and reagents for autophagy detection. When the reagent is used as a single product, it can be detected by flow cytometers as well as fluorescence microscopes. DAPGreen can also be detected using a plate reader. Please select the reagent according to the purpose of your research and the equipment you are using.
| Measurement object | Product Name (Item Code) | Fluorescence properties | Supported Devices | Approximate capacity and number of uses | ||||
|---|---|---|---|---|---|---|---|---|
| Fluorescence microscope | FCM | Plate Reader | ||||||
| Autophagy pathway |
【This product】 |
DALGreen: |
✓ | NA**1 | NA |
1 set |
35 mmdish: 5 dishes |
|
|
DAPRed: |
||||||||
| Autolysosome |
λex=350-450 nm |
✓ | ✓ | NA |
20 nmol |
35 mmdish: 10 dishes |
||
| Autophagosome |
DAPRed - Autophagy Detection (D677) |
λex=500-560 nm λem=690-750 nm |
✓ | NA | NA |
5 nmol |
35 mmdish: 25 dishes |
|
| Autophagosome | λex=425-475 nm λem=500-560 nm *Confocal microscope can excite at 488 nm |
✓ | ✓ | ✓ |
5 nmol |
|||
**1 FCM analysis is available when DALGreen is used by itself.
Note: DALGreen and DAPGreen cannot be co-stained.
Note: The number of uses depends on the staining concentration.
Fluorescent Property
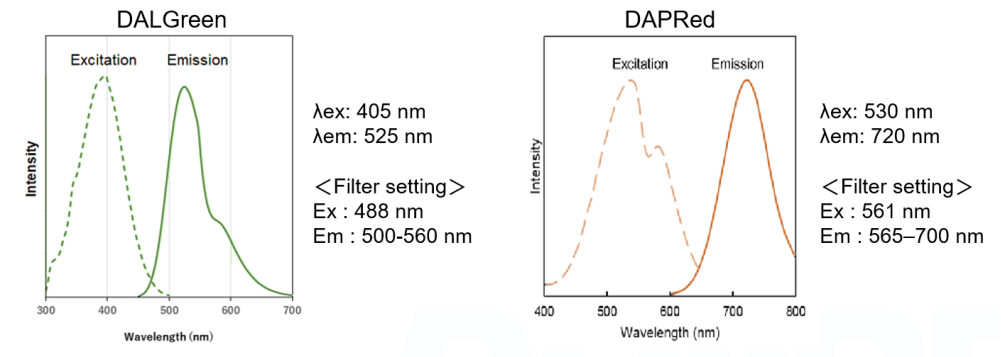
References
| No. | Sample | Publication |
|---|---|---|
| 1) | Cell line (HeLa, MEFs) |
H. Sakurai, et al, "Development of small fluorescent probes for the analysis of autophagy kinetics, iScience, 2023, 26, 107218. |
| 2) | Cell line (HUVECs) |
X. Chen, et al, "Chaiqi decoction ameliorates vascular endothelial injury in metabolic syndrome by upregulating autophagy, Am J Transl Res., 2020, 12(9), 4902-4922. |
| 3) | Cell line (SH-SY5Y) |
C. Oh, et al, "S-Nitrosylation of p62 Inhibits Autophagic Flux to Promote α-Synuclein Secretion and Spread in Parkinson's Disease and Lewy Body Dementia, J Neurosci., 2022, 42(14), 3011-3024. |
Q & A
-
Q
How stable is the DALGreen/DAPRed Working solution?
-
A
DALGreen/DAPRed Working solution cannot be stored. Please prepare fresh Working slution at the time of use.
-
Q
What pathways are involved in autophagy and which states do DALGreen and DAPRed detect?
-
A
It is known that there are two types of autophagy; Atg5-dependent autophagy associated with LC3 modifications, and Atg5-independent autophagy not associated with LC3 modifications*.
DALGreen is taken up by autophagosome membranes and fluoresces in environment when the autophagosomes fused with lysoosmes. This enables the selective detection of autolysosomes with DALGreen.
DAPRed is incorporated into the autophagosome membrane and fluoresces in a hydrophobic environment. DAPRed detects the various states from autophagosomes throughout the autolysosomes.*Reference (Japanese) : Discovery of a New Autophagy Mechanism, Shigeomi Shimizu
https://www.dojindo.co.jp/letterj/160/review/01.html
-
Q
What is the approximate number of uses per kit?
-
A
Please refer to the following for the number of samples that can be measured.
35 mm dish: 5 dishes, μ-Slide 8 well: 5 plates, 96-well Plate: 1 plate
When the final concentrations of DALGreen and DAPRed working solutions are DALGreen : 1.0 µmol/l and DAPRed : 0.2 µmol/l, respectively.
-
Q
Is there anything that I should be careful of when using Bafilomycin A1 for analysis of autolysosome formation inhibition?
-
A
Despite no change in the total fluorescence (area) of the DAPRed fluorescence signals, prolonged Baf. A1 stimulation may cause DAPRed localization to diffuse, resulting in reduced signal visibility on imaging.
Short-term Baf. A1 stimulation does not affect the signal visibility. Please follow the protocol of Experimental Exsample 1 for short-term Baf. A1 stimulation (20 - 40 minutes) after 2 hours of starvation. We have confirmed that the total amount (area) of DAPRed fluorescence signals does not change after prolonged Baf. A1 treatment (8 hours).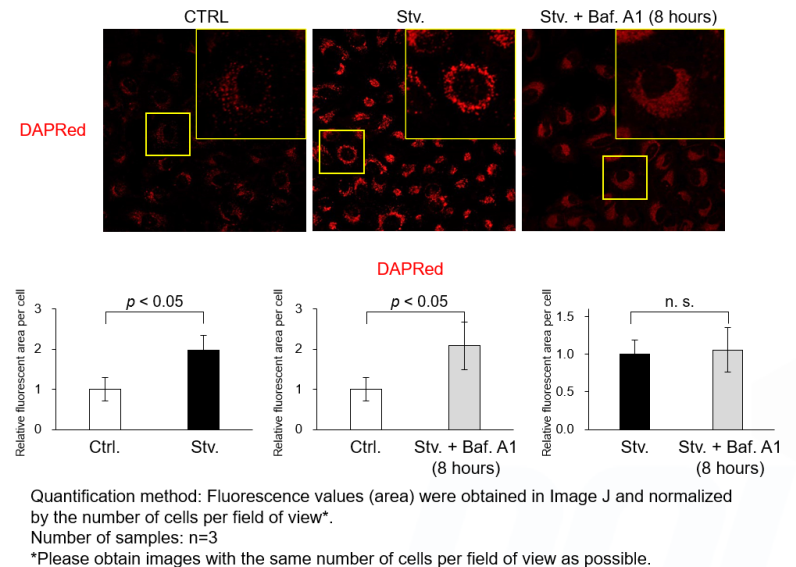
<Experimental Conditions>
CTRL: Normal condition, Stv.: Induction of autophagy, Stv. + Baf. A1: Inhibition of autolysosome
DALGreen filter set: 488 nm (Ex), 490–550 nm (Em)
DAPRed filter set: 561 nm (Ex), 565–700 nm (Em)<Procedure>
1. HeLa cells were seeded (1.0 x 104 cells/well) on a μ-slide 8 well plate (ibidi) and cultured overnight at 37°C in an incubator equilibrated with 95% air and 5% CO2.
2. After washing twice with MEM containing 10% fetal bovine serum, 200 μl of DALGreen/DAPRed working solution (DALGreen: 1 µmol/l, DAPRed: 0.2 µmol/l) and the cells were incubated at 37°C for 30 minutes.
3. The supernatant was discarded, and the cells were washed twice with MEM containing 10% fetal bovine serum.
4. Samples were prepared under the following conditions.• MEM containing 10% fetal bovine serum (200 µl) was added to the well, and the cells were incubated at 37 °C for 8 hours. (Control)
• Amino acid-free medium (FUJIFILM Wako Pure Chemical Industries, Ltd., Catalogue code: 048-33575) (200 μl) was added to the well, and the cells were incubated at 37°C for 8 hours. (Starvation)
• Amino acid-free medium containing Bafilomycin A1 (10,000 times dilution, 200 μl), an inhibitor of lysosomal acidification, was added to the well, and the cells were incubated at 37°C for 8 hours. (Inhibition of autolysosome formation)5. The stained cells were observed under a confocal fluorescence microscope.
-
Q
Is there anything that I should be careful of when using 3-MA for analysis of autophagosome formation inhibition?
-
A
It has been reported that prolonged 3-MA stimulation induces autophagy*. We have confirmed that DAPRed and DALGreen signals were increased compared to control cells in cells pretreated with 3-MA (90 minutes) and incubated with culture medium for 2 hours. Short-term 3-MA stimulation would be better for autophagy inhibition. Please follow the protocol of Experimental Example 2 for short-term 3-MA stimulation (around 20 minutes) after 2 hours of starvation.
*D. Klionsky et. al., "Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition)“, Autophagy, 2016,12(1), 1–222.
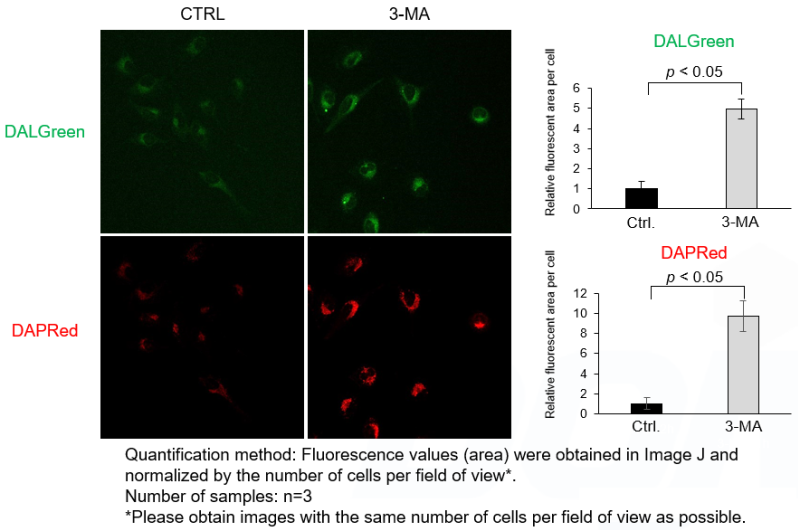
<Experimental Conditions>
CTRL: Normal condition, Stv.: Induction of autophagy, Stv. + 3-MA: Inhibition of autophagosome formation
DALGreen filter set: 488 nm (Ex), 490–550 nm (Em)
DAPRed filter set: 561 nm (Ex), 565–700 nm (Em)<Procedure>
1. HeLa cells were seeded (1.0 x 104 cells/well) on a μ-slide 8 well plate (ibidi) and cultured overnight at 37°C in an incubator equilibrated with 95% air and 5% CO2.
2. The supernatant was discarded, and the cells were washed twice with MEM (containing 10% fetal bovine serum).
3. Samples were prepared under the following conditions.• MEM containing 10% fetal bovine serum (200 µl) was added to the well, and the cells were incubated at 37°C for 1 hour. After washing twice with MEM (containing 10% fetal bovine serum), 200 μl of DALGreen/DAPRed working solution (DALGreen: 1 µmol/l, DAPRed: 0.2 µmol/l) ) was added to the well, and the cells were incubated at 37°C for 30 minutes. (Control)
• MEM containing 10% fetal bovine serum containing 3-MA (10 mmol/l, 200 μl) was added to the well, and the cells were incubated at 37 °C for 1 hour. After washing twice with MEM containing 10% fetal bovine serum, 200 μl of DALGreen/DAPRed working solution (DALGreen: 1 µmol/l, DAPRed: 0.2 µmol/l) containing 3-MA (10 mmol/l) ) was added to the well, and the cells were incubated at 37°C for 30 minutes. (3-MA condition)4. The supernatant was discarded, and the cells were washed twice with MEM containing 10% fetal bovine serum.
5. MEM containing 10% fetal bovine serum (200 µl) was added to the well, and the cells were incubated at 37 °C for 2 hours.
6. The stained cells were observed under a confocal fluorescence microscope.
-
Q
Which autophagy inducers or inhibitors have been analyzed with this kit?
-
A
We have tested the kit in the following autophagy inducers or inhibitors.
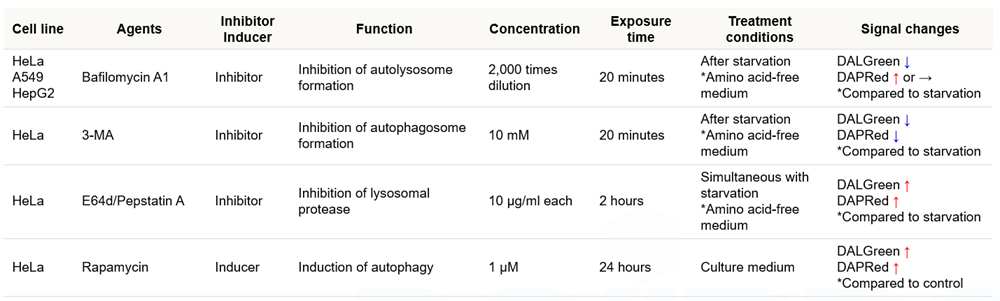
Handling and storage condition
| -20°C, Protect from light, Nitrogen substitution |














