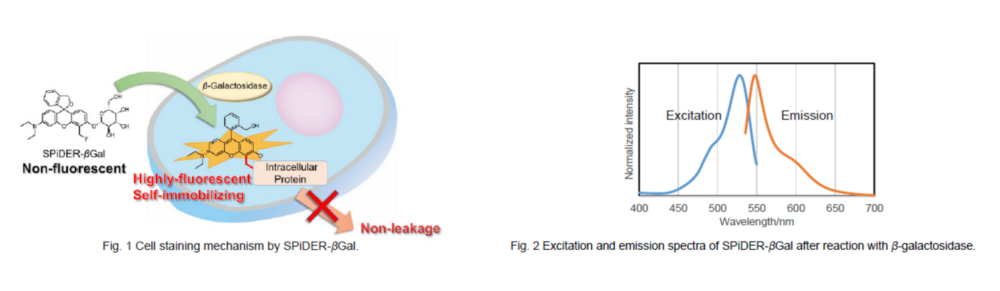
SPiDER-βGal

β-Gal Detection
-
Product codeSG02 SPiDER-βGal
-
CAS No.1824699-57-1
-
Chemical name(2S,3R,4S,5R,6R)-2-{[3'-(Diethylamino)-5'-(fluoromethyl)-3H-spiro(isobenzofuran-1,9'-xanthen)-6'-yl]oxy}-6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol
-
MWC31H34FNO8=567.60
| Unit size | Price | Item Code |
|---|---|---|
| 20 μg x 3 | $622.00 | SG02-10 |
Description
The gene of β-galactosidase from E. coli is widely used as a reporter gene assay marker. Although X-gal is well known reagent to detect β-galactosidase in cell or tissue samples, the assay using these reagents require to fix cells or tissues due to the poor cell-permeability. In addition, so far developed the assay using fluorescence reagents cannot clearly differentiate β-galactosidase-expressed cells or regions.
To overcome these issues, Urano, Kamiya and co-workers have successfully developed SPiDER-βGal. SPiDER-βGal ideally possesses cell-permeability and the ability to retain in intracellular region.1)
By the enzymatic reaction, SPiDER-βGal immediately forms a quinone methide that acts as electrophile when proteins containing nucleophilic functional groups nearby the molecules. By the probe undergoes the reaction with a protein, the conjugates become fluorescent compounds. Thus, SPiDER-βGal allows a single-cell analysis because it does self-immobilizing to the intracellular proteins.
Cellular Senescence Analysis Products
| Product Name | Detection | Sample | Dyes / Fluorescence Properties |
|---|---|---|---|
| Cellular Senescence Detection Kit - SPiDER-βGal | Microscopy or FCM | Living / Fixed cells | SPiDER-βGal Ex: 500–540 nm / Em: 530-570 nm |
| Cellular Senescence Detection Kit - SPiDER Blue | Microscopy, FCM or Plate reader | Fixed cells | SPiDER Blue Ex: 350-450 nm / Em: 400-500 nm |
| SPiDER-βGal | Microscopy | Tissue | SPiDER-βGal Ex: 500–540 nm / Em: 530-570 nm |
| Cellular Senescence Plate Assay Kit - SPiDER-βGal | Plate reader | Living cells | SPiDER-βGal Ex: 500–540 nm / Em: 530-570 nm |
Manual
Technical info
Detection of SA-β-gal in Tissue Samples
A paper on the detection of SA-β-gal in tissue samples from diabetic model mice using SPiDER-βGal has been published.
Quick-frozen tissue was sliced, immersed in 4% paraformaldehyde, and incubated at room temperature for 20 minutes. Then, 20 μmol/L SPiDER-βGal was added to the washed sample in PBS and incubated at 37°C for 1 hour. Finally, the samples were washed with PBS and observed.
For details of the experimental manipulations and data, please refer to this reference.
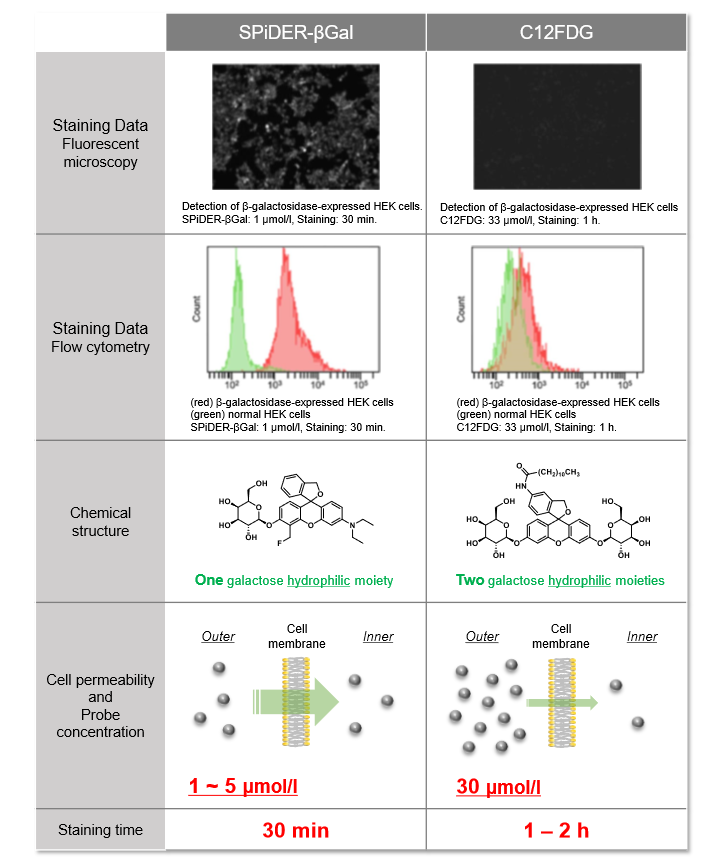
Difference between SPiDER-βGal and C12FDG
Usage Examples:
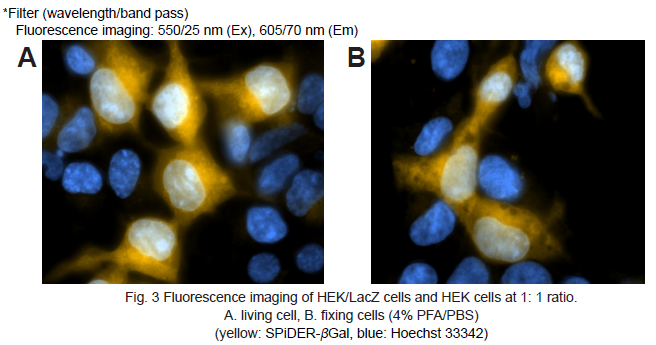
Fluorescence microscopic detection of β-galactosidase-expressed cells
1. HEK cells at 5 × 105 cells/ml (500 μl) and HEK/LacZ cells at 5 × 105 cells/ml (500 μl) were seeded in a 35 mm dish in DMEM (10% fetal bovine serum, 1% penicillin-streptmycin) and cultured overnight in a 5% CO2 incubator at 37oC.
2. The cells were washed with 2 ml of Hanks’ HEPES buffer twice.
3. SPiDER-βGal working solution (2 ml) was added to the culture dish. The cells were then incubated for 15 minutes at 37oC.
4. After the supernatant was removed, the cells were washed Hanks’ HEPES buffer (2 ml) twice.
5. Hanks’ HEPES buffer (2 ml) were added and the cells observed under a fluorescence microscope. (Fig. 3A)
6. After the supernatant was removed, 4% paraformaldehyde (PFA) /PBS solution (2 ml) was added to the culture dish. The cells were then incubated for 15 minutes at room temperature.
7. After 4% PFA/PBS solution was removed, the cells were washed Hanks’ HEPES buffer (2 ml) twice.
8. Hanks’ HEPES buffer (2 ml) were added and the cells observed under a fluorescence microscope. (Fig. 3B)
Flow cytometric detection of β-galactosidase-expressed cells
1. HEK cells at 5 × 105 cells/ml (500 μl) and HEK/LacZ cells at 5 × 105 cells/ml (500 μl) were mixed in a microtube.
2. SPiDER-βGal DMSO stock solution (1 μl) was added to the tube. The cells were then incubated 15 minutes at 37oC .
3. The cells were analyzed under a flow cytometer. (488 nm excitation, 530/30 nm bandpass filter)
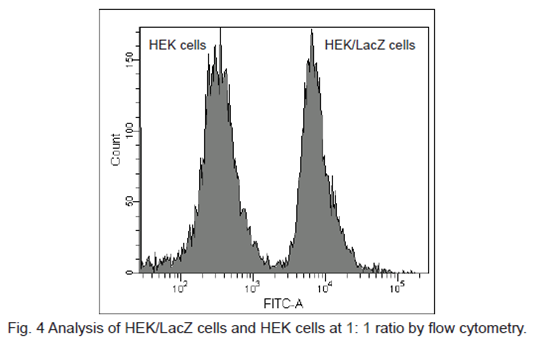
β-galactosidase-expressed cells (HEK/LacZ cells) were clearly differentiate from HEK cells in flow cytometry data analysis.
Live Imaging of Drosophila tissue
Living drosophila tissue was incubated with 10 µmol/l SPiDER-βGal and 16 µmol/l Hoechst 33342 for 20-30 min, observed with confocal microscope.
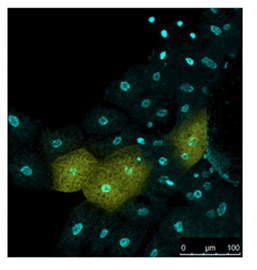
Data was kindly provided by Dr. Y. Urano, at University of Tokyo, Graduate School of Medicine.
Imaging of a Fixed Tissue of a Drosophila
Formalin fixed drosophila generative cells were stained by SPiDER-βGal and DAPI. The β-galactosidase, which is expressed at the nucleus of drosophila generative cells, could be obtained by SPiDER-βGal.
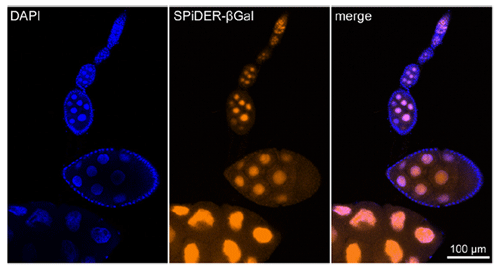
Data was kindly provided by Dr. T. Nakamura, at University of Kumamoto, Institute of Molecular Embryology and Genetics.
Imaging of a Pancreas of a Mouse
SPiDER-βGal and Salmon-Gal were evaluated in the detectability of β-galactosidase on a pancreas of a mouse. SPiDER-βGal was able to detect the difference of the expression point of β-galactosidase more clearly than Salmon-Gal.
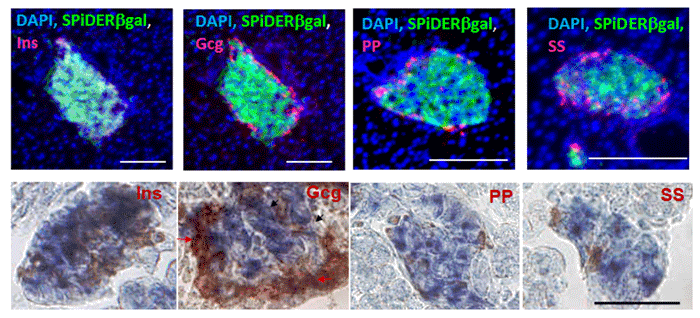
Data was kindly provided by Dr. S. Kume, at Tokyo Institute of Technology, School of Bioscience and Biotechnology.
References
| No. | Sample Type | Instrument | Reference (Link) |
|---|---|---|---|
| 1 | Sample/Tissue (Intestinal stem cells/ Larval wing discs) |
Microscope | T. Doura, M. Kamiya, F. Obata, Y. Yamaguchi, T. Y. Hiyama, T. Matsuda, A. Fukamizu, M. Noda, M. Miura, Y. Urano, "Detection of LacZ-Positive Cells in Living Tissue with Single-Cell Resolution.", Angew Chem Int Ed Engl., 2016, doi: 10.1002/anie.201603328. |
| 2 | Cell (Endocrine cells) |
Microscope | H. Omori, S. Ogaki, D. Sakano, M. Sato, K. Umeda, N. Takeda, N. Nakagata, S. Kume, "Changes in expression of C2cd4c in pancreatic endocrine cells during pancreatic development.", FEBS Lett., 2016, doi: 10.1002/1873-3468.12271. |
| 3 | Cell (SHIN3; SKOV3; OVCAR3) |
Microscope/Flow Cytometer | Y. Nakamura, A. Mochida, T. Nagaya, S. Okuyama, F. Ogata, P. L. Choyke, H. Kobayashi, "A topically-sprayable, activatable fluorescent and retaining probe, SPiDER-βGal for detecting cancer; Advantages of anchoring to cellular proteins after activation", Oncotarget., 2017, doi: 10.18632/oncotarget.17080. |
| 4 | Tissue (Mouse kidney) |
Microscope | S. Lu, S. Liu, A. Wietelmann, B. Kojonazarov, A. Atzberger, C. Tang, R. T. Schermuly, H. J. Gröne, S. Offermanns, "Developmental vascular remodeling defects and postnatal kidney failure in mice lacking Gpr116 (Adgrf5) and Eltd1 (Adgrl4)", PLoS ONE., 2017, 10.1371/journal.pone.0183166. |
| 5 | Tissue (Mouse adipose) |
Microscope | T. Sugizaki, S. Zhu, G. Guo, A. Matsumoto, J. Zhao, M. Endo, H. Horiguchi, J. Morinaga, Z. Tian, T. Kadomatsu, K. Miyata, H. Itoh & Y. Oike, "Treatment of diabetic mice with the SGLT2 inhibitor TA-1887 antagonizes diabetic cachexia and decreases mortality", NPJ Aging Mech Dis., 2017, DOI:10.1038/s41514-017-0012-0. |
| 6 | Cell (VZ/SVZ) |
Flow Cytometer | Y. Nakatani, H. Kiyonari and T. Kondo, "Ecrg4 deficiency results in extended replicative capacity of neural stem cells in a Foxg1-dependent manner", Development., 2019, doi: 10.1242/dev.168120. |
| 7 | Cell (Xenopus oocytes and eggs) |
Microscope | A. A.Tokmakov AA and K. I. Sato, "Activity and intracellular localization of senescence-associated β-galactosidase in aging Xenopus oocytes and eggs.", Exp. Gerontol.., 2019, 119, 157. |
| 8 | Cell (Primary mouse smooth muscle cells) |
Microscope/Flow Cytometer | Y. Han, T. Bedarida, Ye Ding, Q. Wang, P. Song, and M. H. Zou, "β-Hydroxybutyrate Prevents Vascular Senescence through hnRNP A1-Mediated Upregulation of Oct4", Molecular Cell., 2019, 71, 1064–1078. |
| 9 | Cell (Endothelial cells) |
Microscope | A. J. Barinda, K. Ikeda, D. B. Nugroho, D. A. Wardhana, N. Sasaki, S. Honda, R. Urata, S. Matoba, K. Hirata and N. Emoto, "Endothelial progeria induces adipose tissue senescence and impairs insulin sensitivity through senescence associated secretory phenotype", Nat. Commun., 2020, 11, 481. |
| 10 | Tissue (Mouse Muscle Tissue) |
Microscope | Y. Saito, T. Chikenji, T. Matsumura, M. Nakano, and M. Fujimiya, "Exercise enhances skeletal muscle regeneration by promoting senescence in fibro-adipogenic progenitors", Nat. Commun., 2020, doi:10.1038/s41467-020-14734-x. |
| 11 | Cell (HUVEC Cells) |
Flow Cytometer | M. Suda, I. Shimizu, G. Katsuumi, Y. Yoshida, Y. Hayashi, R. Ikegami, N. Matsumoto, Y. Yoshida, R. Mikawa, A. Katayama, J. Wada, M. Seki, Y. Suzuki, A. Iwama, H. Nakagami, A. Nagasawa, R. Morishita, M. Sugimoto, S. Okuda, M. Tsuchida, K. Ozaki, M. Nakanishi-Matsui and T. Minamino, "Senolytic vaccination improves normal and pathological age-related phenotypes and increases lifespan in progeroid mice", Nature Aging, 2021, doi:10.1038/s43587-021-00151-2. |
| 12 | Cell (MG63 cells) |
Microscope | H. Nakashima, M. Yasunaga, M. Yoshida, M. Yamaguchi, S. Takahashi, H. Kajiya, S. Tamaoki, and J. Ohno, "Low Concentration of Etoposide Induces Enhanced Osteogenesis in MG63 Cells via Pin1 Activation", J. Hard Tissue Biol., 2021, doi:10.2485/jhtb.30.175. |
| 13 | Cell (Mouse muscle cells) |
Microscope | V. Moiseeva, A. Cisneros, V. Sica, O. Deryagin, Y. Lai, S. Jung, E. Andres, J. An, J. Segales, L. Ortet, V. Lukesova, G. Volple, A. Benguria, A. Dopazo, S. Aznar-Benitah, Y. Urano, A. d. Sol, M. A. Esteban, Y. Ohkawa, A. L. Serrano, E. Perdiguer & P. Munoz-Canoves, "Senescence atlas reveals an aged-like inflamed niche that blunts muscle regeneration", Nature, 2022, doi:10.1038/s41586-022-05535-x. |
Q & A
-
Q
Is there comparison data with X-Gal? What is an advantage over X-Gal?
-
A
SPiDER-βGal allows stain of a sample with a shorter time and without fixing the sample.
Please check the comparison data below. There is comparison data between SPiDER-βGal and X-Gal for tissue staining.
【Mouse Kidney】
Refer to Figure S14 and 15 on the Supporting Information.
【Mouse Salivary Gland】
Refer to Figure S14 and 15 on the Supporting Information.
【Mouse Brain】
Refer to Figure S16 and 17 on the Supporting Information.
-
Q
What is an advantage over commercially available β-Galactosidase detection probe?
-
A
SPiDER-βGal has higher cell permeability and intracellular retention than any other commercially available probe, which allows stain of a sample with a shorter time and lower concentration for mammalian cells. Please check the Selection Guide for more details.
-
Q
Can I use SPiDER-βGal for tissue staining?
-
A
Yes, SPiDER-βGal can be used for tissue staining. We have experimental data with the following samples:
【Drosophila Tissue】
Refer to Figure S9, 12 and 13 on the Supporting Information.
【Mouse Kidney】
Refer to Figure S14 and 15 on the Supporting Information.
【Mouse Salivary Gland】
Refer to Figure S14 and 15 on the Supporting Information.
【Mouse Brain】
Refer to Figure S16 on the Supporting Information.
-
Q
Can I fix a sample after staining with SPiDER-βGal?
-
A
Yes, the sample stained with SPiDER-βGal can be fixed with 4% PFA or methanol.
-
Q
Can I stain a fixed sample with SPiDER-βGal?
-
A
Yes, SPiDER-βGal can be used for the fixed sample. We have experimental data for HEK cell, drosophila and mouse tissue. However, since fixation causes lower β-galactosidase activity, please optimize the condition for fixation.
-
Q
How many time can I assay with 1 kit (20 μg×3) ?
-
A
If the final concentration is prepared at 1 μmol/l for cell staining, you can do approximately fifty 35mm dishes, fifty 8-chamber plates and ten 96- well plates.
-
Q
Is there information regarding cytotoxicity?
-
A
Cytotoxicity of SPiDER-βGal to HEK-lacZ(+) cells and HEK cells was measured with Cell Counting Kit-8.
【Cytotoxicity assay with HEK-lacZ(+) cells and HEK cells】
Refer to Figure S8 on the Supporting Information.
-
Q
How long is the Working solution stable?
-
A
You can’t store the working solution. Please prepare the working solution prior to use.
-
Q
What buffer can I use for preparing the Working solution?
-
A
You can use PBS, Hanks’ HEPES and HBSS etc.
-
Q
What is the recommended filter?
-
A
Fluorescence microscopy: Ex.550/25 nm, Em.605/70 nm
Flow Cytometry: Ex.488 nm, Em.530/30 nm
Handling and storage condition
| Appearance: | Red to red purple solid |
|---|---|
| Purity (HPLC): | ≧ 85.0 % |
| NMR spectrum: | Authentic |
| Fluorescence spectrum: | To pass test |
| 0-5°C |