-Nucleostain- DNA Damage Quantification Kit -AP Site Counting-
DNA Damage Detection
-
Product codeDK02 -Nucleostain- DNA Damage Quantification Kit -AP Site Counting-
| Unit size | Price | Item Code |
|---|---|---|
| 20 samples | $486.00 | DK02-12 |
| 20 samples | ・ARP-DNA Standard Solution ・ARP Solution ・DNA Binding Solution ・Washing Buffer ・HRP-Streptavidin ・TE Buffer ・Substrate Solution ・Filtration Tube ・96-well Microplate/U Bottom |
each 250 μl × 1 250 μl × 1 10 ml × 1 × 1 25 μl × 1 40 ml × 1 10 ml × 1 20 tubes × 1 |
|---|
Product Description
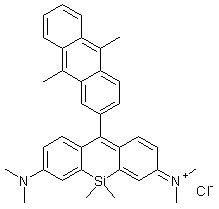
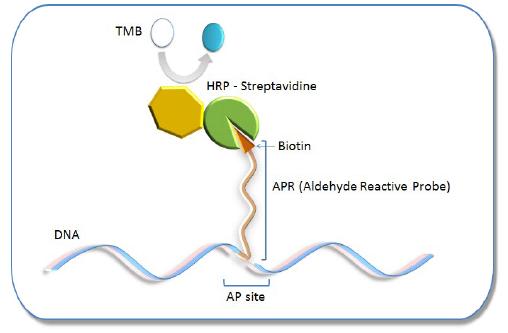
Oxidative damage to DNA is a result of its interaction with reactive oxygen species (ROS), in particular, the hydroxy radical. Hydroxy radicals, which are produced from superoxide anion and hydrogen peroxide by the Fenton reaction, produce multiple modifications in DNA. Oxidative attacks by hydroxy radicals on the deoxyribose moiety will lead to the release of free bases from DNA, generating strand breaks with various sugar modifications and simple abasic sites (AP sites). In fact, AP sites are one of the major types of damage generated by ROS. Aldehyde Reactive Probe (ARP; N Eaminooxymethylcarbonylhydrazin-D-biotin) reacts specifically with an aldehyde group present on the open ring form of the AP sites (Fig. 1). This reaction makes it possible to detect DNA modifications that result in the formation of an aldehyde group. After treatment with excess ARP reagent, all of the AP sites on DNA are tagged with a biotin residue. These biotin-tagged AP sites can be quantified using the avidin-biotin assay, followed by colorimetric detection with either peroxidase or alkaline phosphatase conjugated to the avidin. DNA Damage Quantification Kit contains all the necessary solutions for detecting between 1 to 40 AP sites per 1 x 105 base pairs.
AP site Detection Principle
Mechanism of ARP Tagging at an Abasic Site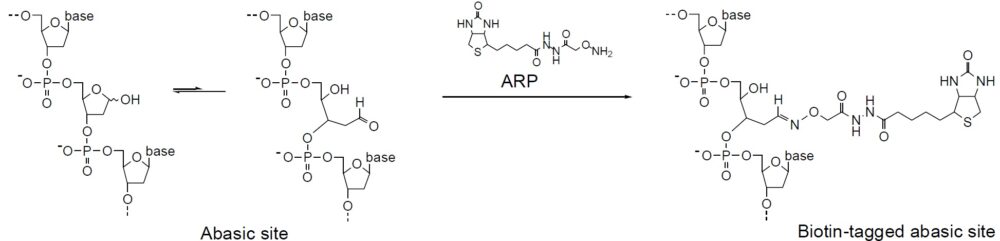
| Developer | Dojindo Molecular Technologies, Inc. |
|---|
Manual
Technical info
How to Prepare a Calibration Curve
1. Calculate the average O.D. of each ARP-DNA standard solution.
2. Subtract the blank O.D. from the average O.D.a)
3. Plot the O.D. corresponding to the number of AP sites of the standard solution. X-axis is the number of AP sites and Y-axis is the O.D.
4. Determine the number of AP sites in the sample using this calibration curve.
a) The blank O.D. is about 0.04-0.06 and the O.D. of the 40 ARP DNA standard solution is about 0.8-1.0. The O.D. value depends on HRPStreptavidin activity.
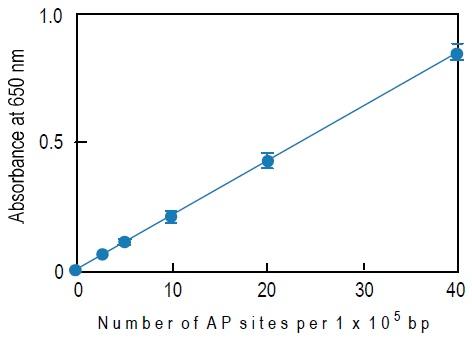
Fig. 3 Typical calibration curve of DNA Damage Quantification Kit
References
1) A. Sancar and G. B. Sancar, "DNA Repair Enzymes", Annu. Rev. Biochem., 1988, 57, 29.
2) T. Lindahl and B. Nyberg, "Rate of Depurination of Native Deoxyribonucleic Acid", Biochemistry, 1972, 11, 3610.
3) M. Liuzzi and M. Talpaert-Borle, "A New Approach to the Study of the Base-excision Repair Pathway Using Methoxyamine", J. Biol. Chem., 1985, 260, 5252.
4) M. Weinfeld, M. Liuzzi and M. C. Paterson, "Response of Phage T4 Polynucleotide Kinase Toward Dinucleotides Containing Apurinic Sites: Design of a 32P-postlabeling Assay for Apurinic Sites in DNA", Biochemistry, 1990, 29, 1737.
5) B. X. Chen, K. Kubo, H. Ide, B. F. Erlanger, S. S. Wallace and Y. W. Kow, "Properties of a Monoclonal Antibody for the Detection of Abasic Sites, a Common DNA Lesion", Mutat. Res., 1992, 273, 253.
6) J. A. Gralnick and D. M. Downs, "The YggX Protein of Salmonella enterica Is Involoved in Fe(II) Trafficking and Minimizes the DNA Damage Cause by Hydroxyl Radicals:Residue CYS-7 is Essential for YggX Function", J. Biol. Chem., 2003, 278, 20708.
7) J. W. Pippin, R. Durvasula, A. Petermann, K. Hiromura, W. G. Couser and S. J. Shankland, "DNA Damage is a Novel Response to Sublytic Complement C5b-9 Induced Injury in Podocytes", J. Clin. Invest., 2003, 111, 877.
8) S. Watanabe, T. Ichimura, N. Fujita, S. Tsuruzono, I. Ohki, M. Shirakawa, M. Kawasuji and M. Nakao, "Methylated DNA-binding Domain 1 and Methylpurine DNA Glycosylase Link Transcriptional Repression and DNA Repair in Chromatin", Proc. Natl. Acad. Sci. USA, 2003, 100, 12859.
9) M. Endres, M. Ahmadi, I. Kruman, D. Biniszkiewicz, A. Meisel and K. Gertz, "Folate Deficiency Increases Postischemic Brain Injury", Stroke, 2005, 36, 321.
10) J.-M. Li, M. Mogi, K. Tsukuda, H. Tomochika, J. Iwanami, L.-J. Min, C. Nahmias, M. Iwai and M. Horiuchi, "Angiotensin II-Induced Neural Differentiation via Angiotensin II Type 2 (AT2) Receptor-MMS2 Cascade Involving Interaction between AT2 Receptor-Interacting Protein and Src Homology 2 Domain-Containing Protein-Tyrosine Phosphatase 1", Mol. Endocrinolo., 2007, 21(2):499.
11) D. R. McNeill and D. M. Wilson III, "A Dominant-Negative Form of the Major Human Abasic Endonuclease Enhances Cellular Sensitivity to Laboratory and Clinical DNA-Damaging Agents", Mol. Cancer Res., 2007, 5(1), 61.
Q & A
-
Q
Can I use single-stranded DNA or RNA?
-
A
No, you cannot use this kit to determine the number of abasic sites in single-stranded DNA or RNA. The O.D. reading of single-stranded DNA will be nearly twice that of double-stranded DNA because of the binding efficiency on the microplate.
-
Q
How should genomic DNA be stored?
-
A
Prepare a DNA pellet and store at -20°C or -80°C if the DNA cannot be labeled with ARP immediately after isolation. After ARP labeling, the sample can be stored at 4°C in TE Buffer for several months.
-
Q
How should I prepare the DNA?
-
A
You can use general protocols or commercially available DNA isolation kits. Between 2 to 4 abasic sites per 1 x 105 base pairs will be created during the DNA isolation process. Therefore, use the same isolation method to prepare each DNA sample.
-
Q
What should I do if the sample DNA concentration is less than 100 μg per ml?
-
A
You can either use a filtration tube to concentrate your sample DNA or ethanol precipitation to recover DNA as a pellet and then re-dissolve it to prepare a 100 μg per ml solution.
-
Q
What should I do if the sample DNA is less than 1 μg?
-
A
Add the same volume of ARP Solution and follow the manual. The recovery of the ARP-labeled DNA may be lower than the usual reactions, so measure the ARP-labeled DNA solution. The average recovery rate of the 0.5 μg DNA and 0.25 μg DNA is 70% and 50%, respectively.
Handling and storage condition
| 0-5°C | |
|
Danger / harmful symbol mark |

|
|---|---|