ExoSparkler Exosome Membrane Labeling Kit-Red

Exosome Membrane Fluorescent Staining
- Observe exosome dynamics accurately without extracellular aggregation
- Cover steps from fluorescence labeling to purification
-
Product codeEX02 ExoSparkler Exosome Membrane Labeling Kit-Red
| Unit size | Price | Item Code |
|---|---|---|
| 5 samples | $300.00 | EX02-10 |
*Protein amount : 1-10μg/ sample, Particle count : 10 to 100 x 108 /samples
(As purified exosome using ultracentrifugation)
| 5 samples | ・Mem Dye-Red ・Filtration tube |
×1 ×5 |
|---|
Description
Product Description
Exosomes are a form of extracellular vesicle (EV), which could contribute to malignant transformation and the metastasis of cancer. Consequently, intercellular communication via exosomes is attracting considerable interest in the scientific community. To shed light on such communication, labeling techniques based on fluorescent dyes have been used. Fluorescent dyes that label the cellular membrane are commonly used for exosome labeling because the lipid bilayer in exosomes is a suitable labeling target. ExoSparkler series can be used for staining of purified exosomal membrane or protein, which allows imaging of labeled exosomes taken up by cells.
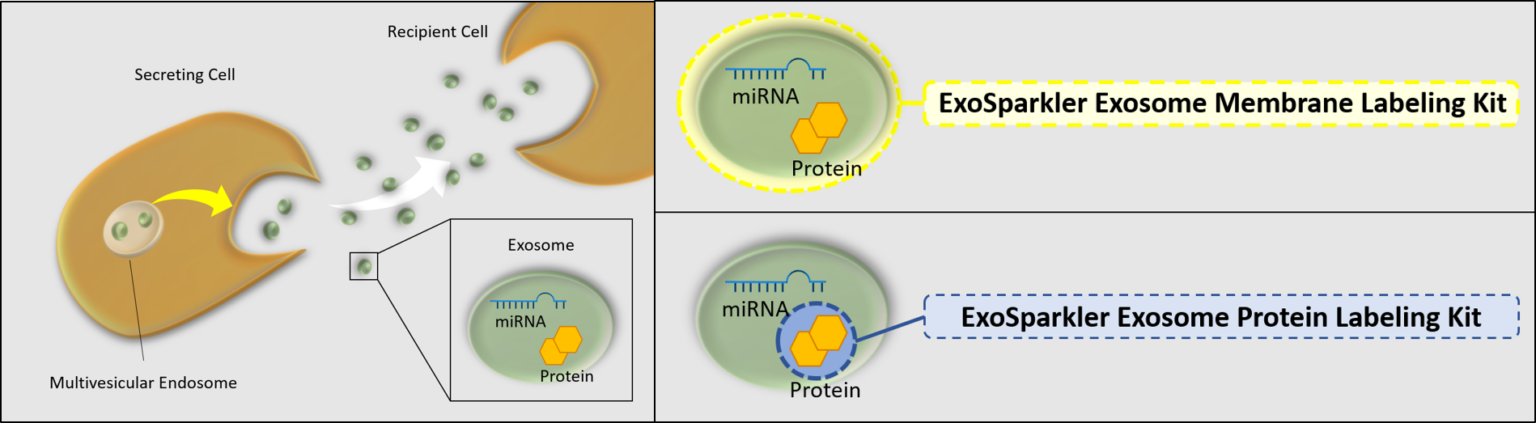
Manual
Technical info
Commonly used exosomal membrane dye can cause dye aggregation, exhibiting fluorescent spots that are not derived from exosomes. These dyes can also change the functional properties of exosomes while increasing the background imaging.1,2
The dyes used in ExoSparkler series (Mem Dye-Green, Red, and Deep Red) do not cause aggregation and have little influence on properties of exosomes, allowing a more accurate observation of exosome dynamics.
1) Mehdi Dehghani et al., “Exosome labeling by lipophilic dye PKH26 results in significant increase in vesicle size”.bioRxiv., 2019, doi:10.1101/532028.
2) Pužar Dominkuš P et al., “PKH26 labeling of extracellular vesicles: Characterization and cellular internalization of contaminating PKH26 nanoparticles.” Biochim Biophys Acta Biomembr., 2018, doi: 10.1016/j.bbamem.2018.03.013.
ExoSparkler series does not allow extracellular aggregation
Exosomes stained with ExoSparkler’s Mem Dye-Deep Red or an alternative product (green or red) were added to each well containing HeLa cells. The labeled exosomes taken into HeLa cells were observed by fluorescent microscopy. As a result, extracellular fluorescent spots suspected of dye aggregations were seen in each well containing the exosomes stained with the alternative product (green or red).

Experimental conditions
Exosomes were purified by ultracentrifugation (10μg exosome protein) and stained with each dye. Labeled exosomes were added to HeLa cells (1.25×104 cells), and the cells were incubated for 24 hours. Cells were washed, and immunofluorescence images showing labeled exosomes were observed.
Detection conditions
Mem Dye-Deep Red(Purple): Ex 640nm/Em 640-760nm
Alternative Product “P” (Green): Ex 561nm/Em 560-620nm
Alternative Product “P” (Red): Ex 640nm/Em 650-700nm
Mem Dye-Deep Red and Product “P” (Green and Red) in aqueous solution were analyzed by NTA (nanoparticle tracking analysis) to investigate the generation of aggregates. No aggregation was observed in the experiments with Mem Dyes, although Product “P” (Green and Red) produced dye-to-dye aggregates (100–500 nm size).
Instrument: LM10-HSBFT 14 (Nanosight)
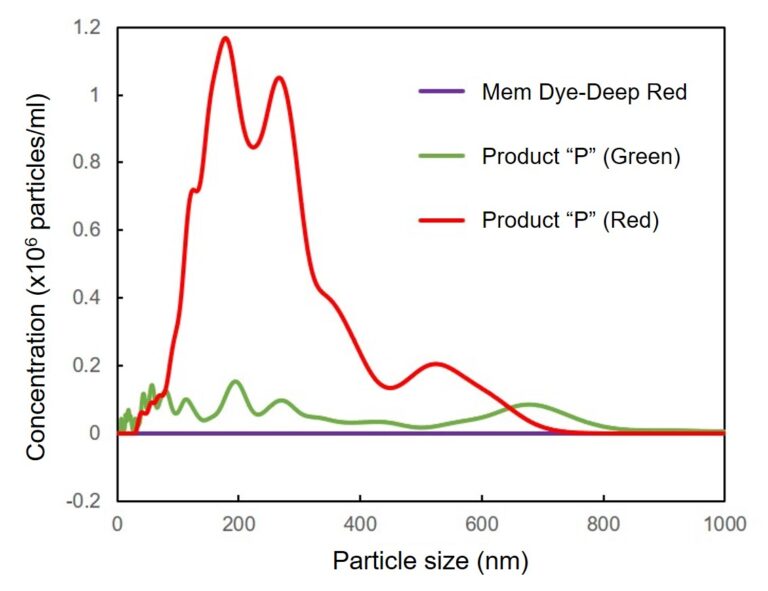
Change the particle size of Mem Dye-solution and Product “P” solution
In Mem Dye-Green, Red, the aggregation of the dye was not confirmed as in Mem Dye-Deep Red.
Mem Dyes have little effect on exosome properties
NTA (nanoparticle tracking analysis) and zeta potential were measured to determine the changes in exosomes of dye-stained with Mem Dye-Deep Red or Product “P” (green or red) or unstained exosomes.
As a result, the Mem-Dye series (green, red, deep red) had little effect on exosome properties.
Effect of the dyes on the particle size of the exosomes
Exosomes were stained with Mem Dye-series (green, red, deep red) and Product “P” (green and red) at a dye concentration of 10 μmol/L in DMSO, the NTA (nanoparticle tracking analysis) of the stained exosomes (as 10 µg protein) was measured.
As a result, Mem Dyes-series did not change number and particle size of the exosomes (bottom left). Conversely, the Product “P” stained exosomes showed the significant changes of particle size and population of the exosomes (bottom right).
Instrument: LM10-HSBFT 14 (Nanosight)
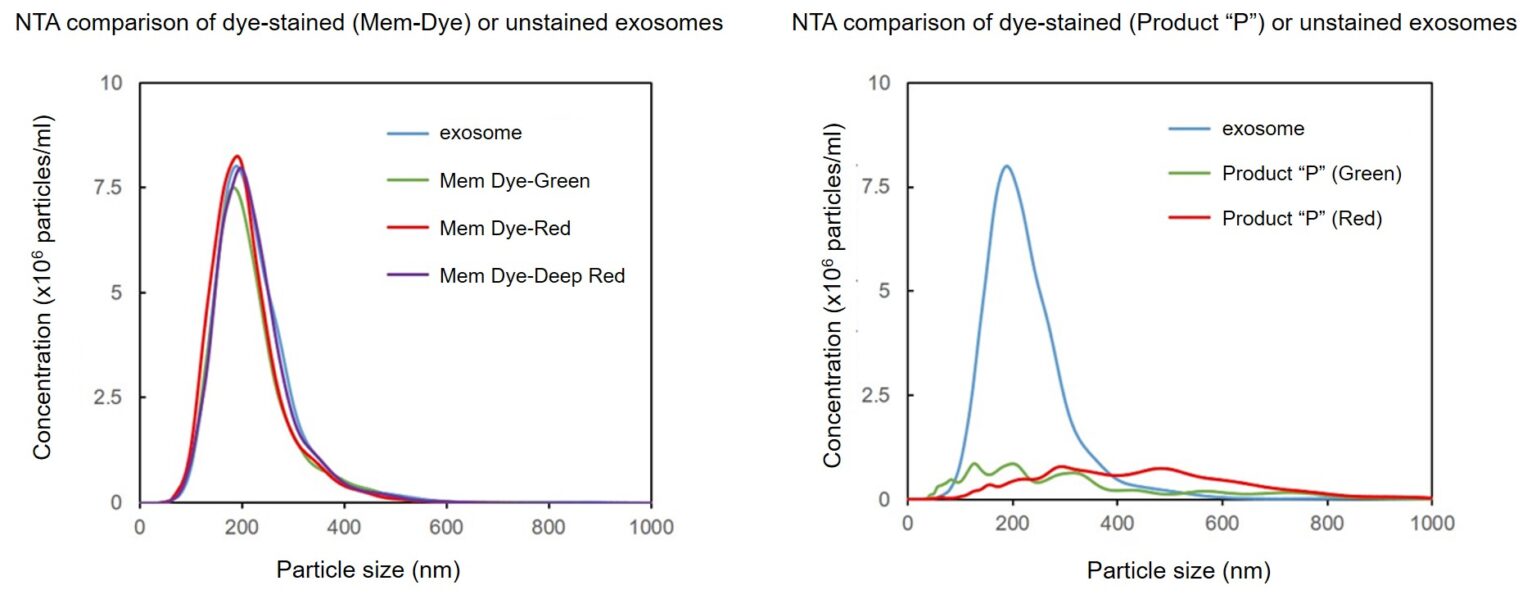
Effect of the dyes on the zeta potentials of the exosomes
Exosomes were stained with Mem Dye-series (green, red, crimson) and Product “P” (green and red) at a dye concentration of 10 μmol/L in DMSO, the zeta potentials of the stained exosomes (as 10 µg protein) was measured.
As a result, product “P”-stained exosomes have lower zeta potential than Mem Dye-stained.
Instrument: Zetasizer Nano ZSP (Malvern Panalytical)
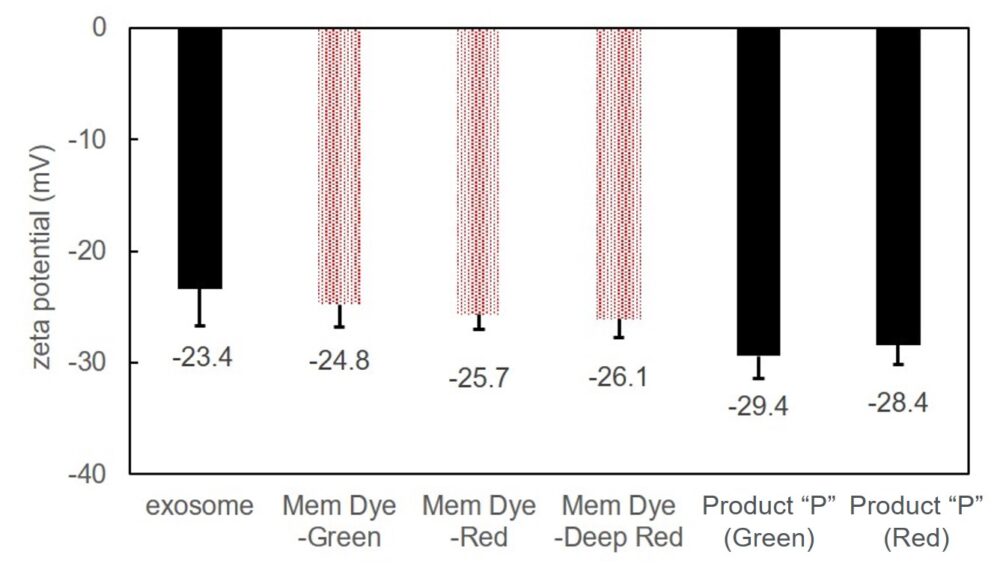
Zeta potentials comparison of dye-stained (Mem-Dye/Product “P”) or unstained exosomes
References) Takashi Shimomura et al., “New Lipophilic Fluorescent Dyes for Exosome Labeling: Monitoring of Cellular Uptake of Exosomes”.bioRxiv., 2020, doi:10.1101/2020.02.02.931295.
Our ExoSparkler Exosome Membrane Labelling Kits provide everything from fluorescence labeling to purification
ExoSparkler series contains filtration tubes available for the removal of dyes unreacted after fluorescence labeling, as well as an optimized protocol for labeling exosomes. Our ExoSparkler series makes it possible to prepare fluorescence labeling of exosomes using the simple procedure.
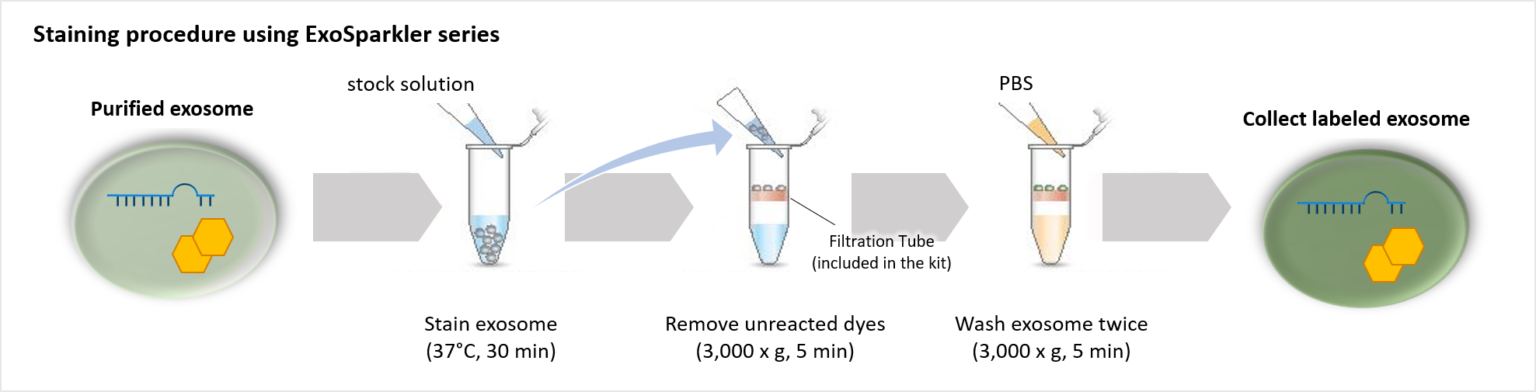
Comparison of purification methods (removal of unlabeled dyes)
The filtration tubes used to remove unlabeled dyes in this kit can purify exosomes at a higher recovery rate than gel filtration methods.

For the effectiveness of purification using filtration tubes, please see Q&A.
(The filter is colored in the purification after the labeling, Have unlabeled dyes been removed?)
Observe the time-dependent changes in exosomes localization
Exosomes purified by ultracentrifugation (10 µg as protein amount) were stained with Mem Dye-Deep Red (Exosome Membrane Fluorescence Labeling Kit) and added to HeLa cells (1.25×104 cells) stained with lysosome staining dye. The fluorescence images were observed after 1 h and 4 h incubation.
As a result, it was confirmed that the fluorescence puncta (purple) of Mem Dye-Deep Red overlapped with the localization of lysosomes (green) over time (white), and that the localization of exosomes changed in a time-dependent manner.
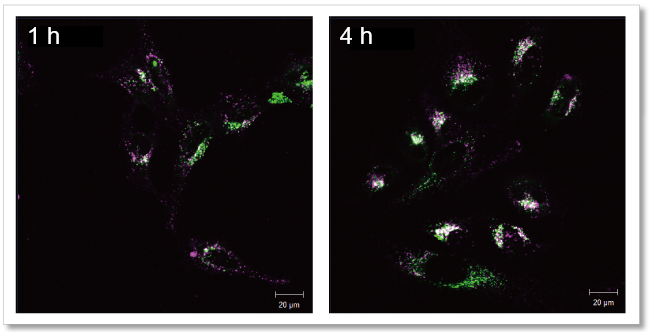
Mem Dye-Deep Red (Purple): Ex 640 nm/Em 640-760 nm
Lysosome staining dye: Ex 488 nm/Em 490-540 nm
Visualization of EVs uptake via endocytic pathway
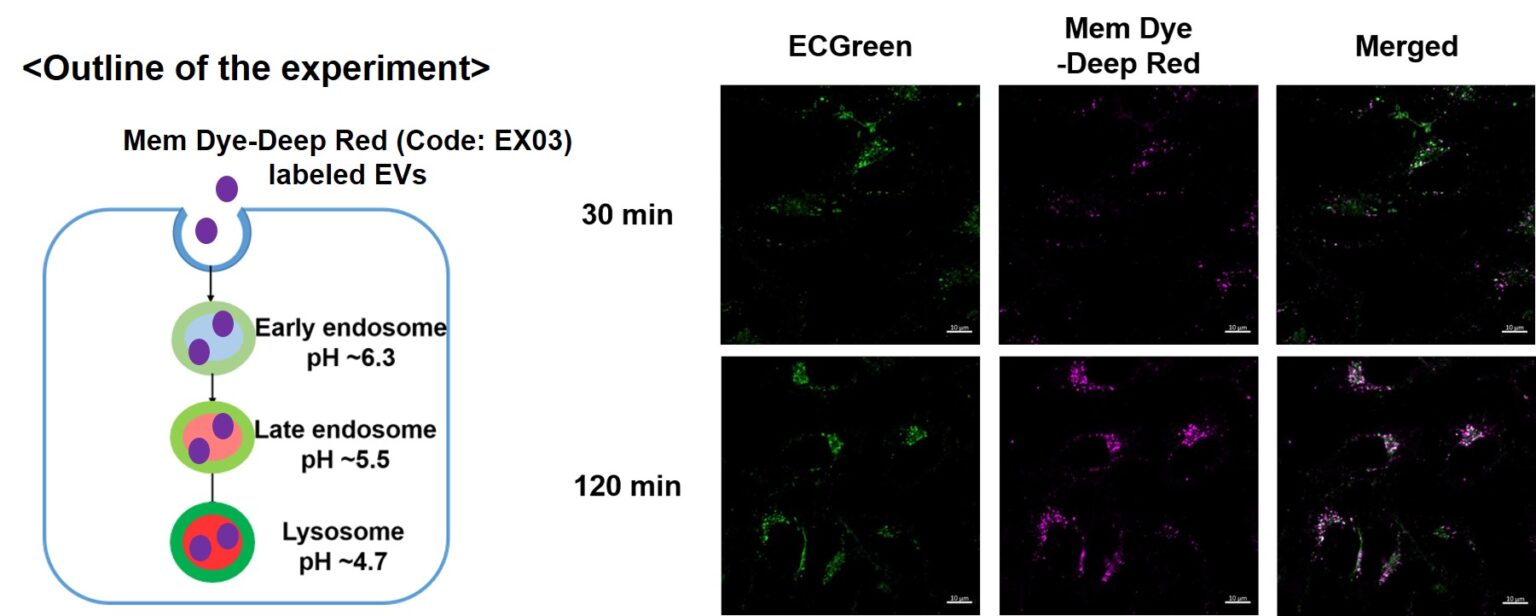
Mem Dye-labeled EVs are internalized via endocytosis:
HeLa cells were incubated with 10 μmol/L ECGreen (Code: E296) for 30 min. Then, Mem Dye-Deep Red labeled EVs (quantified as 10 µg of protein) were added to HeLa cells. After 30 or 120 min incubation, the cells were washed and observed under a fluorescence microscope (Scale Bar: 10 µm).
ExoSparkler series product comparison
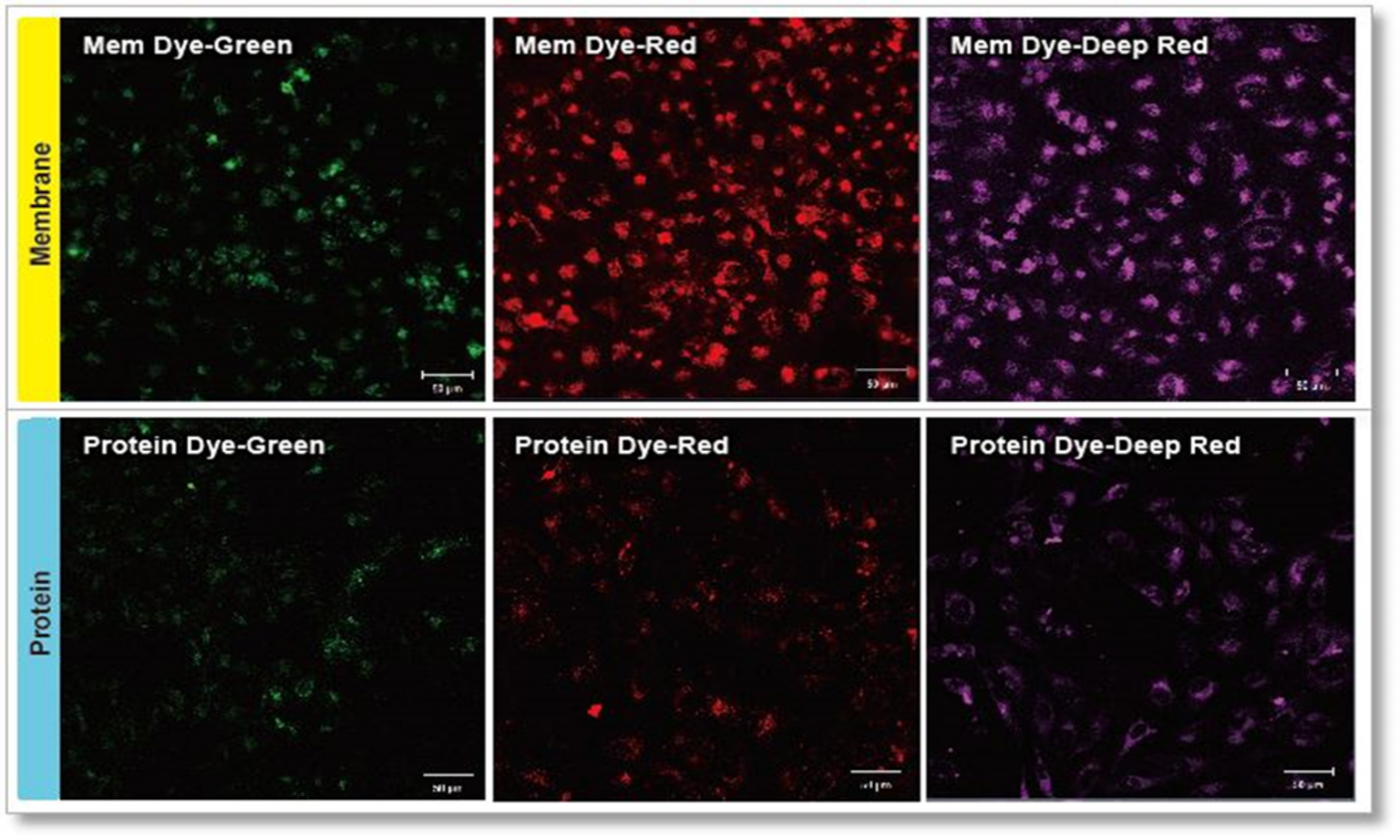
Experimental conditions
Exosomes were purified by ultracentrifugation (10 μg exosome protein) and stained with each dye. Labeled exosomes were added to HeLa cells (1.25×104 cells), and the cells were incubated for 24 hours. Cells were washed, and immunofluorescence images showing labeled exosomes were observed.
Detection conditions
Green: Ex 488nm/Em 490-540nm
Red: Ex 561nm/Em 570-640nm
Deep Red: Ex 640nm/Em 640-760nm
References
| No. | Sample | Instrument | Reference (Link) |
|---|---|---|---|
| 1) | Cell (RAW264.7) |
Fluorescent Microscope | 1) R.Kawata, S.Oda, Y. Koya, H.Kajiyama, T. Yokoi, "Macrophage-derived extracellular vesicles regulate concanavalin A-induced hepatitis by suppressing macrophage cytokine production", Toxicology, 2020, https://doi.org/10.1016/j.tox.2020.152544. |
| 2) | Cell (ADSCs) |
Fluorescent Microscope | A. Shimoda R. Miura, H. Tateno, N. Seo, H. Shiku, S. Sawada, Y. Sasaki and K. Akiyoshi, "Assessment of Surface Glycan Diversity on Extracellular Vesicles by Lectin Microarray and Glycoengineering Strategies for Drug Delivery Applications", Small Methods, 2021, doi:10.1002/smtd.202100785. |
| 3) | HEK293T cells-derived | Fluorescent Microscope | Y. Matsuki, T. Yanagawa, H. Sumiyoshi, J. Yasuda, S. Nakao, M. Goto, T. Shibata-Seki, T. Akaike, Y. Inagaki, "Modification of exosomes with carbonate apatite and a glycan polymer improves transduction efficiency and target cell selectivity", 2021, doi:10.1016/j.bbrc.2021.10.063. |
| 4) | Colorectal Cancer Cells-derived | Fluorescent Microscope | H. Takakura, T. Nakao, T. Narita, M. Horinaka, Y. Nakao-Ise, T. Yamamoto, Y. Iizumi, M. Watanabe, Y. Sowa, K. Oda, N. Mori, T. Sakai, and M. Mutho, "Citrus limon L.-Derived Nanovesicles Show an Inhibitory Effect on Cell Growth in p53-Inactivated Colorectal Cancer Cells via the Macropinocytosis Pathway ", 2022, doi:10.3390/biomedicines10061352. |
Q & A
-
Q
Are there recommended purification methods for exsome?
-
A
We recommended the exosomes prepared by ultracentrifuge. We also have experience labeling the exosomes purified by immunoprecipitation and magnetic beads.
You can also use the ExoIsolator Exosome Isolation Kit (EX10), which has been proven the recovery rate equivalent to that of the ultracentrifugal method.
In addition, exosomes prepared by the polymer-based precipitation method are not applicable for the following reasons.
1) The polymer may clog the filtration tube.
2) The polymer may interact with exosomes and change the membrane potential of exosomes. If the membrane potential changes, the intracellular kinetics of labeled exosomes may change when they are used in cellular uptake assays.
-
Q
Some dyes remains on the filte after purification. Have unlabeled dyed been removed?
-
A
Unlabeled dye is likely to be retained inside the filter, but we have confirmed that the unlabeled dye is not remaining on top of the filter. Please refer to the following experiment:
① We stained the 10 µg exosomes (as protein amount) purified by ultracentrifugation.
② We also prepared only using buffer as a control
3. The recovered solution is added to HeLa cells (1.25 x 104 cells), and the fluorescence image was observed 4 hours later.
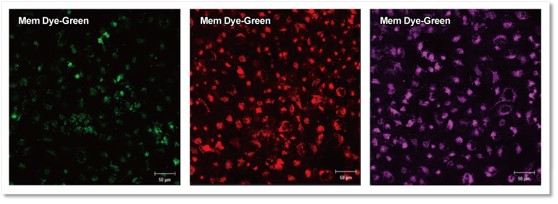
As a result, it was confirmed that no bright fluorescent particles (unlabled dyes) were observed in the cells using the recovered solution with the only buffer.
① Exosome + Buffer
② Only Buffer

Fluorescent images at 4 h incubation
Detection conditions
Green:Ex 488 nm / Em 490 – 540 nm
Red :Ex 561 nm / Em 570 – 640 nm
Deep Red:Ex 640 nm / Em 640 – 760 nm
-
Q
Can I store the exosomes after it has been stained?
-
A
We can not recommend to store the exosomes after it has been stained.
Please use the stained exosomes as soon as possible.
-
Q
Which media have been used in exosome uptake experiments?
-
A
We had evaluated the exosome uptake experiments using MEM (Minimum Essential Medium) and DMEM (Dulbecco’s Modified Eagle’s Medium). We can not recommend to use the serum-free medium because this kit has been optimized to observe in serum-containing medium.
-
Q
What is the amount of exosomes that can be labeled per sample?
-
A
Isolated exosomes (ultracentrifugation method), protein: 1-10 μg/sample, number of particles: 10-100x 108 particles/sample.
-
Q
If there are contaminants other than exosomes, are they labeled by ExoSparkler, too?
-
A
Yes, if there are contaminants in the sample, such as proteins or polymers*, they will be labeled by ExoSparkler too. Therefore, please prepare purified exosomes.
For details on the purification method, please refer to the FAQ "Are there recommended purification methods for exosomes?"*Polymers may remain depending on the purification method.
Handling and storage condition
| 0-5°C, Protect from light、Protect from moisture |















