Cellular Senescence Detection Kit - SPiDER-βGal
Senescence Cell Detection
- Quantify SA-βgal
- Applicable for Living Cell and Fixed Cell
- Staining time 30 min.
-
Product codeSG04 Cellular Senescence Detection Kit - SPiDER-βGal
| Unit size | Price | Item Code |
|---|---|---|
| 1 plate | $260.00 | SG04-01 |
| 3 plates | $590.00 | SG04-03 |
| 10 plates | $1293.00 | SG04-10 |
The same amount of SG03-10 is with SG04-03.
*Difference between SG03 and SG04 is the only amount of reagent.
| 1 plate | ・SPiDER-βGal ・Bafilomycin A1 |
×1 ×1 |
|---|---|---|
| 3 plates | ・SPiDER-βGal ・Bafilomycin A1 |
×3 ×3 |
| 10 plates | ・SPiDER-βGal ・Bafilomycin A1 |
×10 ×10 |
Description
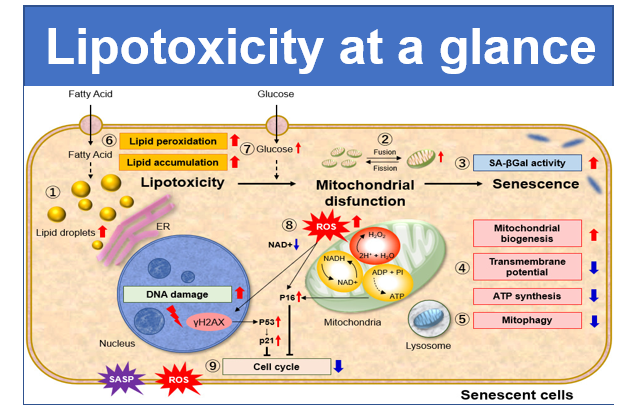
DNA damages of the normal cells are caused by repeated cell division and oxidative stress. Cellular Senescence, a state of irreversible growth arrest, can be triggered in order to prevent DNA-damaged cells from growing. Senescence-associated β-galactosidase (SA-β-gal), which is overexpressed in senescent cells, has been widely used as a marker of cellular senescence. Although X-gal is a well known reagent to detect SA-β-gal, these are following disadvantages: 1) requirement of fixed cells due to the poor cell-permeability, 2) low quantitative capability because of the difficulty of the determination of visual difference between stained cells and not stained cells, 3) requirement of a long time of staining.
Cellular Senescence Detection Kit – SPiDER-βGal allows to detect SA-β-gal with high sensitivity and ease of use. SPiDER-βGal is a new reagent to detect β-galactosidase which possesses a high cell-permeability and a high retentivity inside cells. SA-β-gal are detected specifically not only in living cells but also fixed cells by using a reagent (Bafilomycin A1) to inhibit endogenous β-galactosidase activity. Therefore, SPiDER-βGal can be applied to quantitative analysis by flow cytometry.
Recent work from Dr. Kim et al. at Mayo Clinic used our Cellular Senescence Detection Kit – SPiDER-βGal to evaluate cellular senescence in endothelial cells. They did staining of SA-βGal in cells in atherosclerotic renal artery stenosis (ARAS) and co-stained the cells with CD31 which is a marker of endothelilal cells. They showed that ARAS + Elamipretide* treatment slightly improved endothelial cell senescence. Unlike commercial available probes used for detection of β-Galactosidase, SPiDER-βGal contained in the kit possesses high intracellular retention. The key feature of this product is that it can be used to co-stain SA-β-Gal and other markers. Our kit is a useful tool for cellular senescence research.
*Elamipretide: mitochondria-targeted peptide
For more information on data, please refer to the publication below:
S. R. Kim, A. Eirin, X. Zhang, A. Lerman and L. O. Lerman, “Mitochondrial Protection Partly Mitigates Kidney Cellular Senescence in Swine Atherosclerotic Renal Artery Stenosis.”, Cell. Physiol. Biochem. ., 2019, 52, 617.
Cellular Senescence Analysis Products
| Product Name | Detection | Sample | Dyes / Fluorescence Properties |
|---|---|---|---|
| Cellular Senescence Detection Kit - SPiDER-βGal | Microscopy or FCM | Living / Fixed cells | SPiDER-βGal Ex: 500–540 nm / Em: 530-570 nm |
| Cellular Senescence Detection Kit - SPiDER Blue | Microscopy, FCM or Plate reader | Fixed cells | SPiDER Blue Ex: 350-450 nm / Em: 400-500 nm |
| SPiDER-βGal | Microscopy | Tissue | SPiDER-βGal Ex: 500–540 nm / Em: 530-570 nm |
| Cellular Senescence Plate Assay Kit - SPiDER-βGal | Plate reader | Living cells | SPiDER-βGal Ex: 500–540 nm / Em: 530-570 nm |
Manual
Technical info
Principle
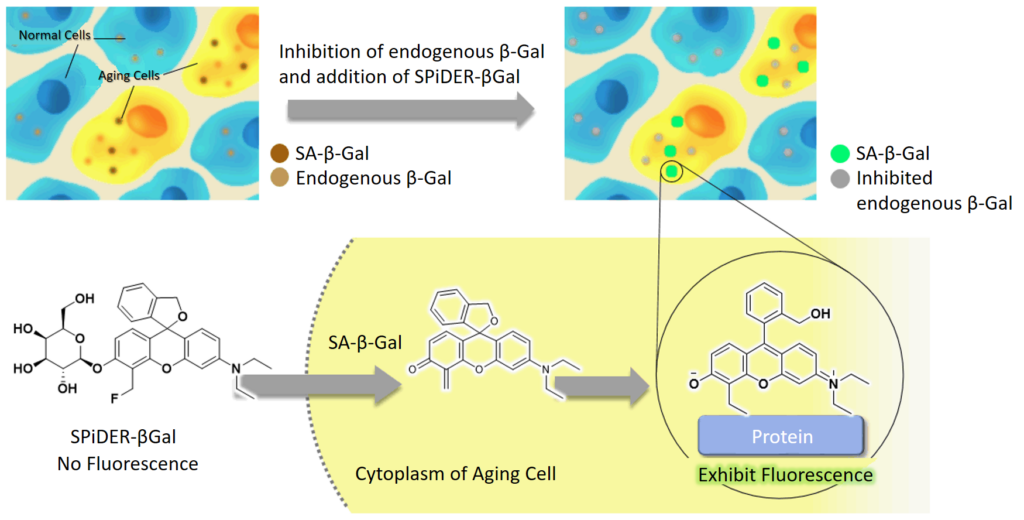
SPiDER-βGal, the β-galactosidase detection reagent in this kit, is cell membrane permeable. Thus, SPiDER-βGal is adapted to live cells without membrane permeabilization or fixation procedures. After SPiDER-βGal permeates the cell membrane, it reacts with SA-βGal to fluoresce and form covalent bonds with neighbouring proteins. Furthermore, the addition of bafilomycin A1 contained in this kit before SPiDER-βGal reaction suppresses the activity of endogenous β-galactosidase in living cells, allowing fluorescence detection of SA-βGal in a background suppressed state.
Fluorescence property of SPiDER-βGal reacted with β-galactosidase
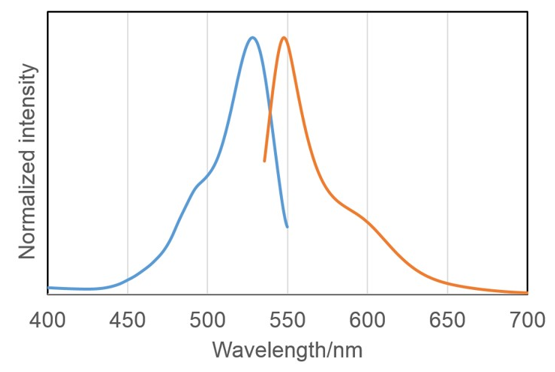
| <Tested experimental conditions> | λex | λem |
| Confocal microscope | 488 | 500-600 |
| Epifluorescence microscope | 545/25 | 605/70 |
| Flow cytometry | 488 | 530/30 |
Comparison of SG04 with existing methods
| Product name | Living cells | Fixed cells | Intracellular retention | Detection | Suitable devices | Staining time | Supplementary information |
|---|---|---|---|---|---|---|---|
|
Cellular Senescence Detection Kit |
++ | ++ | +++ | Fluorescence | Fluorescence microscope Flow cytometry |
30 min |
・Sensitivity is high, allowing short staining times and quantitative analysis ・Applcable for multiple staining with other senescence markers |
| ++ | NA | NA | Fluorescence | Plate reader | 30 min |
・SA-βGal activity is quantified by simple addition of the reagents to a 96-well microplate. ・Applcable for multiple sample analysis |
|
| C12FDG | + | + | + | Fluorescence | Fluorescence microscope Flow cytometry |
1-2 hours | ・The staining time of C12FDG is longer than that of SPiDER βGal due to its lower cell membrane permeability. |
| X-Gal | NA | ++ | ++ | Colorimetry | Microscope | Overnight | ・Objective measurement is difficult due to visual counting. |
Simple Procedure
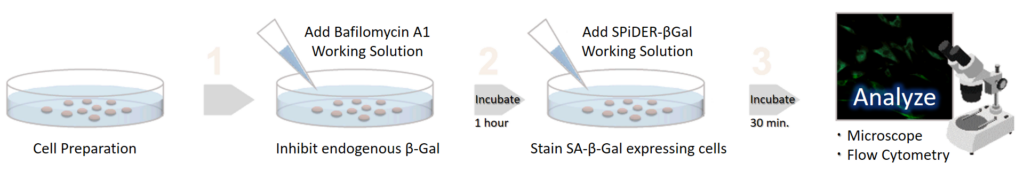
SPiDER-βGal is applicable to living cells and fixed cells
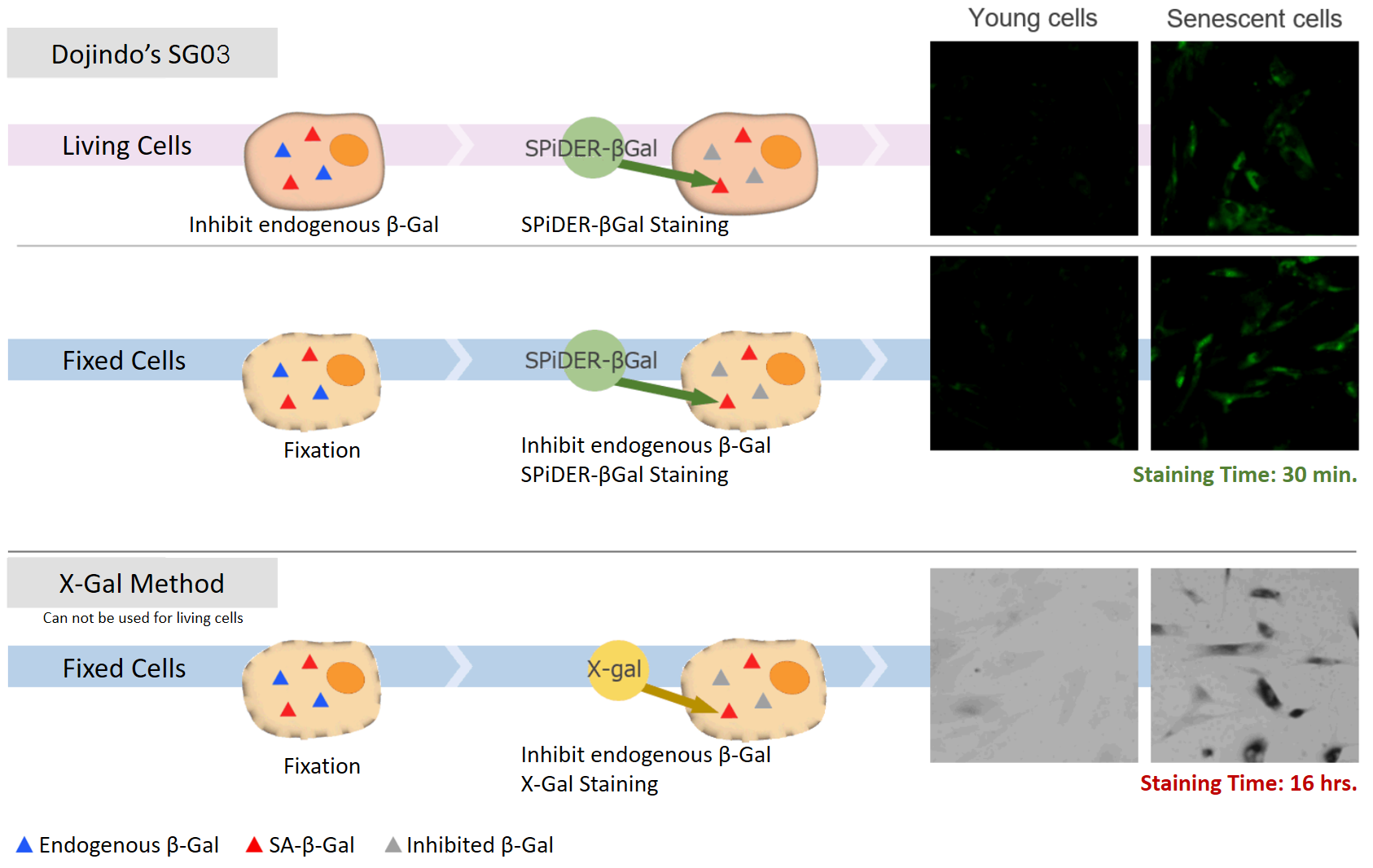
Applicable for SA-βGal quantification

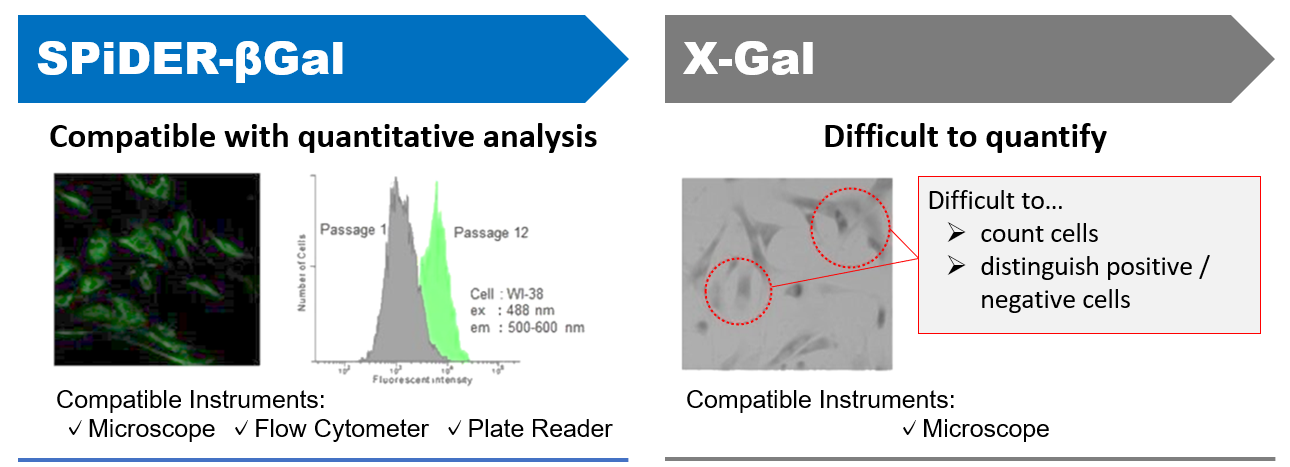
For plate reader detection, please refer Cellular Senescence Plate Assay Kit - SPiDER-βGal.
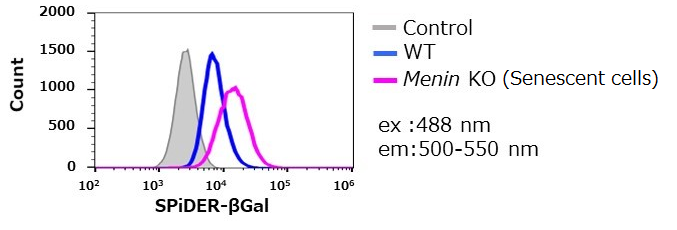
SA-β-gal detection for T cells (floating cells)
The research group by professor Masakatsu Yamashita at Ehime University Graduate School of Medicine has shown that a protein called Menin controls T cell exhaustion, aging, and maintains normal immune function.
By using this kit, they confirmed that SG03 has the ability to detect induced cell senescence by stimulating TCR (T cell receptor) in the presence of interleukin 2 (IL-II) in naive CD8+ T cells that are knocked out Menin.
Staining Conditions
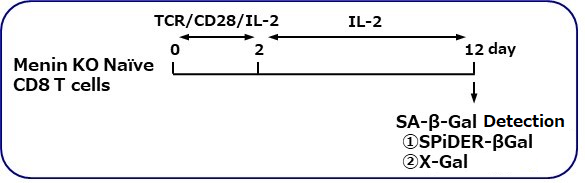
① SPiDER-βGal method
② X-gal method
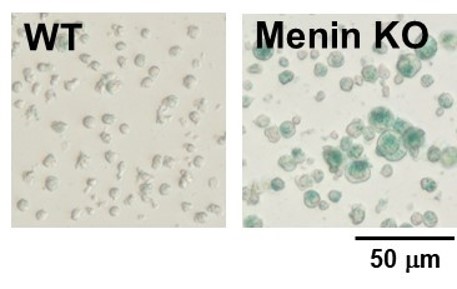
*Data was kindly provided by Masakatsu Yamashita, at Ehime University Graduate School of Medicine
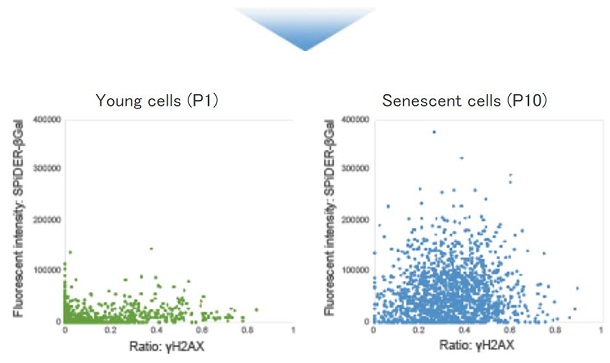
Quantification with confocal quantitative image cytometer
Quantitative analysis of senescent cells using a confocal quantitative image cytometer (Yokogawa Electric Corporation, CQ1).
■ Quantitative analysis of senescent cells co-stained with SA-βGal and other senescence markers.
Live WI-38 cells were stained with SPiDER-βGal. After then, the stained cells were fixed, permeabilized and immunostained for the DNA damage marker γ-H2AX. Quantitative analysis was performed in confocal quantitative imaging cytometers.
.jpg)
|
|
■ Analysis of fluorescently stained living cells.
In the conventional method of X-gal, SA-β-gal-positive cells are counted under microscope and calculate the percent of the senescent cells by compared with total cells. The SA-β-gal-positive cells were stained with this kit and total cells were stained with Hoechst 33342. Stained cells were analyzed using confocal quantitative image cytometer CQ1.
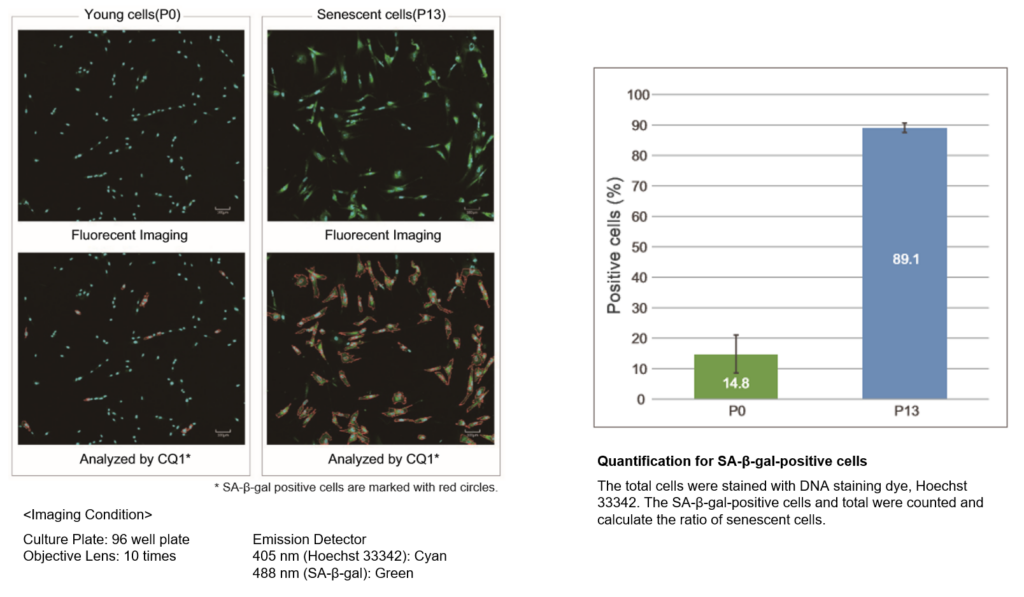
The difference of SA-β-gal-positive cells ratio were shown in WI-38 cells depending on the number of passage. The data was quickly analysed with the confocal quantitative image cytometer compared with the manually counting procedure with X-gal staining method.
SA-β-gal detection for tissue
In this published article, SA-β-gal was detected using SPiDER-βGal on tissue samples from a diabetic model mouse.
Note: SPiDER-βGal [Code: SG02] is used in this staining.
After slicing the frozen tissue, it was fixed with 4% paraformaldehyde for 20 minutes at room temperature. Then it was washed with PBS and observed.
For details of the experimental operation and data, refer to Reference 2) below.
Markers of Senescent Cells
Co-staining of SA- β-gal and DNA Damage marker in WI-38 cells
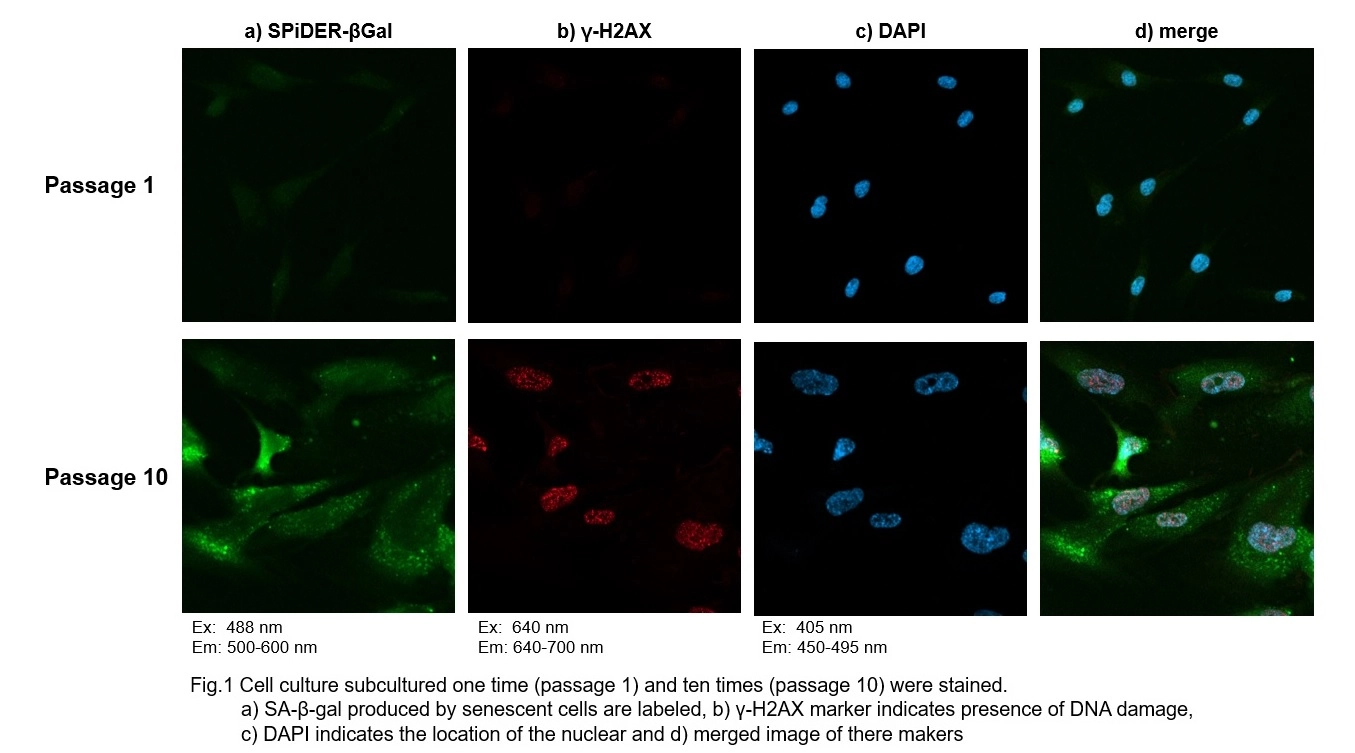
Procedure:
1. Passage 1 and 10 of WI-38 were used. The procedure was followed as the manual within the kit.
2. Add 4% PFA/PBS to the cells and incubate for 15 minutes at room temperature
3. Wash the cells 3 times with PBS
4. Add 0.1% Triton X-100/PBS to cells and incubate for 30 minutes at room temperature
5. Wash the cells 3 times with PBS
6. Add 1% BSA/PBS to the cells and incubate for 1 hour at the room temperature
7. Add anti- γ-H2AX antibody (rabbit) diluted with 1% BSA/PBS to the cells and incubate at 4℃ overnight
8. Wash the cells 3 times with PBS
9. Add Anti- rabbit secondary antibody (Alexa Fluor 647) diluted with 1% BSA/PBS to the cells and incubate at room temperature for 2 hours
10. Wash cells 3 times with PBS
11. Add 2 μg/ml DAPI (code: D523) diluted with PBS to the cells and incubate for 10 minutes at room temperature
12. Wash cells 3 times with PBS and observe under a confocal microscope
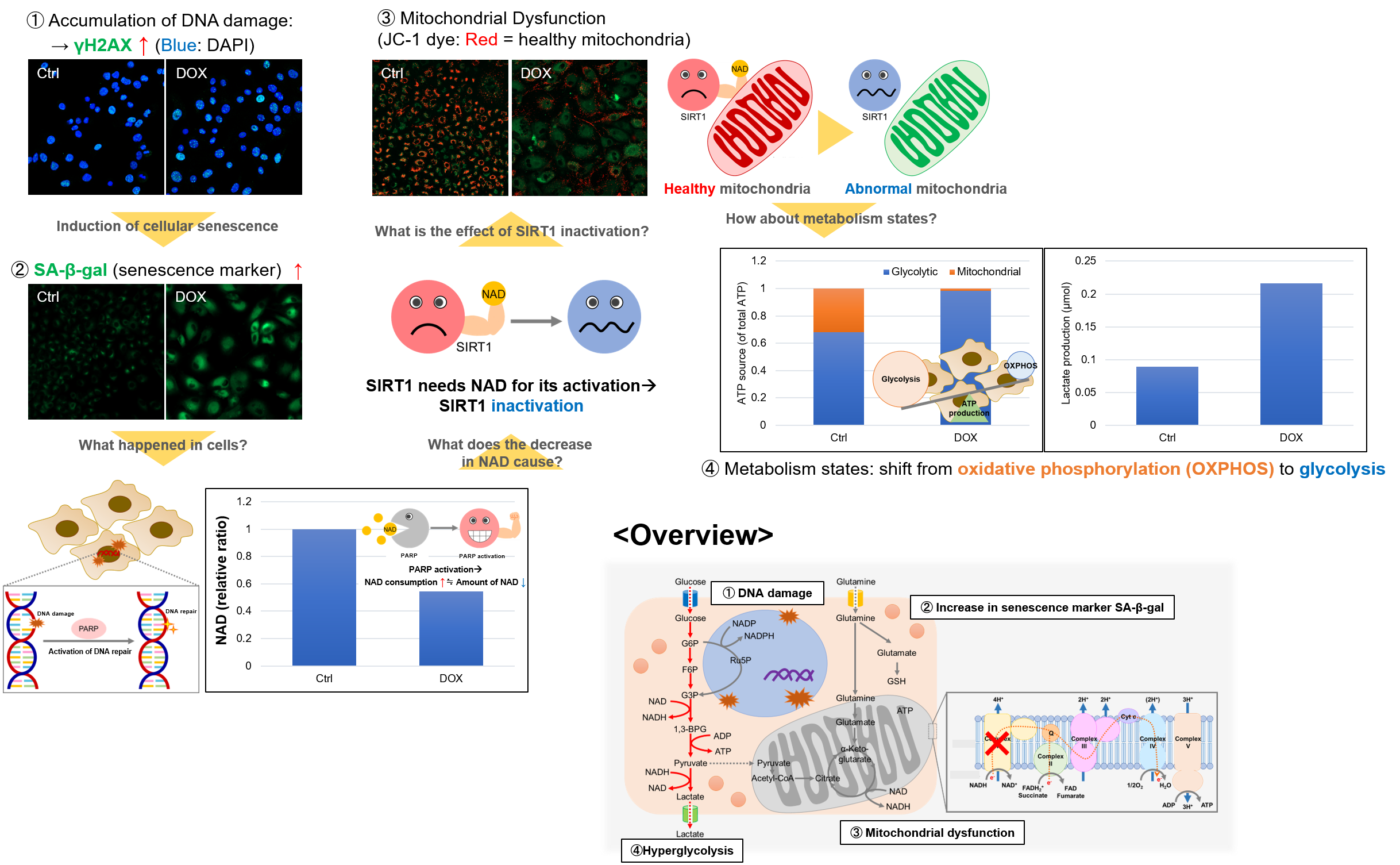
Experiment Example: Cellular Senescence with Metabolic shift
NAD(+) levels decline during the aging process, causing defects in nuclear and mitochondrial functions and resulting in many age-associated pathologies*. Here, we try to redemonstrate this phenomenon in the doxorubicin (DOX)-induced cellular senescence model with a comprehensive analysis of our products.
*S. Imai, et al., Trends Cell Biol, 2014, 24, 464-471
Related Products
| Category | Product | Unit | SKU |
|---|---|---|---|
|
Cellular Metabolism |
100 tests |
N509-10 |
|
| Lactate Assay Kit-WST | 50 / 200 tests | L256-10 / L256-20 | |
| Glycolysis/OXPHOS Assay Kit NEW | 50 tests | G270-10 | |
|
Cellular Senescence Detection |
1 / 3 / 10 plates |
SG04-01 / SG04-03 / SG04-10 |
|
|
Mitochondrial Membrane Potential |
10 set |
MT09-10 |
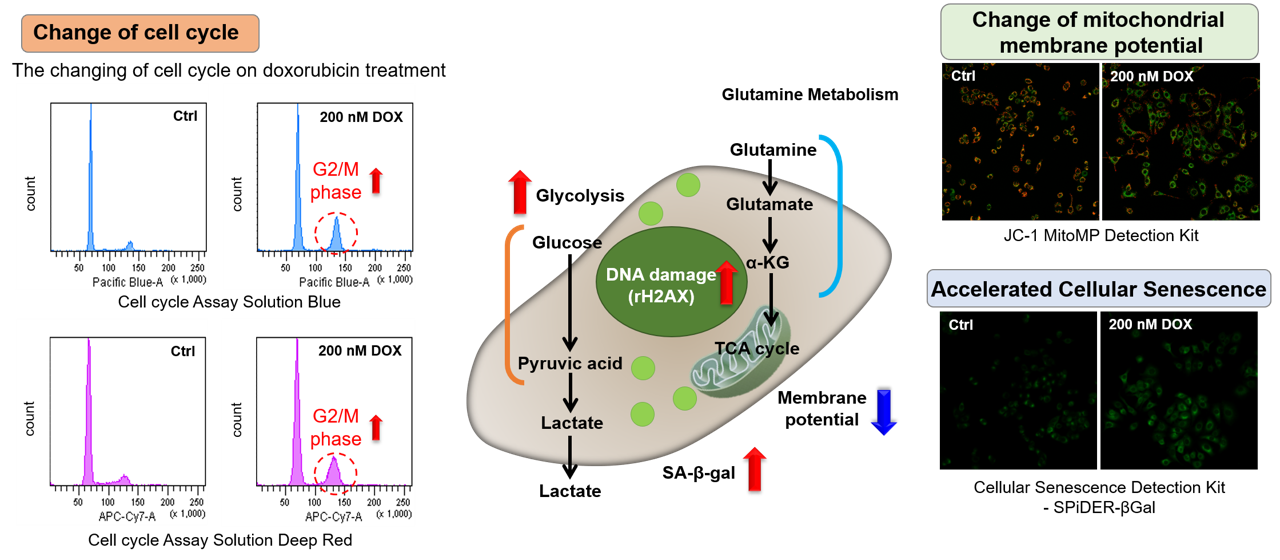
Association between cellular senescence and cell cycle
Doxorubicin (DOX) acts to inhibit cell proliferation during G2/M phases of the cell cycle and induces cellular senescence. After adding DOX to A549 cells, higher histogram peaks for the G2/M phase (Cell Cycle Assay Solution Blue and Deep Red), induces cellular senescence (Cellular Senescence Detection Kit - SPiDER-βGal), and the differences in mitochondrial membrane potential (JC-1 MitoMP Detection Kit) were observed.
Supplementary information on technology and products used
-

Cell Cycle Measurement Reagent
Cell Cycle Assay Solution Blue
-
Mitochondrial Membrane Potential Detection Kit
JC-1 MitoMP Detection Kit
Conditions for senescence induction and measuring device
| Cell type (tissue) and Conditions for senescence induction | Measuring device | Product code | Reference |
|---|---|---|---|
|
・MRC cells |
Fluorescence microscope |
SG04 |
A. Park, I. Tsunoda and O. Yoshie, "Heat shock protein 27 promotes cell cycle progression by down-regulating E2F transcription factor 4 and retinoblastoma family protein p130", J. Biol. Chem.., 2018, doi: 10.1074/jbc.RA118.003310 . |
|
・A549 cells |
Fluorescence microscope, Flow cytometry |
SG04 |
R. Tanino, Y. Tsubata, N. Harashima, M. Harada and T. Isobe, "Novel drug-resistance mechanisms of pemetrexed-treated non-small cell lung cancer", Oncotarget., 2018, 9, (24), 16807. |
|
・NHDF cells |
Fluorescence microscope |
SG04 |
Y. Kitahiro, A. Koike, A. Sonoki, M. Muto, K. Ozaki and M. Shibano. , "Anti-inflammatory activities of Ophiopogonis Radix on hydrogen peroxide-induced cellular senescence of normal human dermal fibroblasts.", J Nat Med ., 2018, 72, 905. |
|
高脂肪試料およびマウス脂肪組織 |
Fluorescence microscope |
SG02 |
T. Sugizaki, S. Zhu, G. Guo, A. Matsumoto, J. Zhao, M. Endo, H. Horiguchi, J. Morinaga, Z. Tian, T. Kadomatsu, K. Miyata, H. Itoh & Y. Oike, "Treatment of diabetic mice with the SGLT2 inhibitor TA-1887 antagonizes diabetic cachexia and decreases mortality", Nature Partner Journal:Aging and Mechanisms of Disease., doi:10.1038/s41514-017-0012-0. |
|
Intestinal epithelial organoid (6 and 78 weeks old mice) |
Fluorescence microscope |
SG04 |
R. Uchida, Y. Saito, K. Nogami, Y. Kajiyama, Y. Suzuki, Y. Kawase, T. Nakaoka, T. Muramatsu, M. Kimura and H. Saito , "Epigenetic silencing of Lgr5 induces senescence of intestinal epithelial organoids during the process of aging", NPJ Aging Mech Dis., 2018,doi:10.1038/s41514-018-0031-5. |
Co-staining of SA- β-gal and DNA Damage marker in fixed WI-38 cells
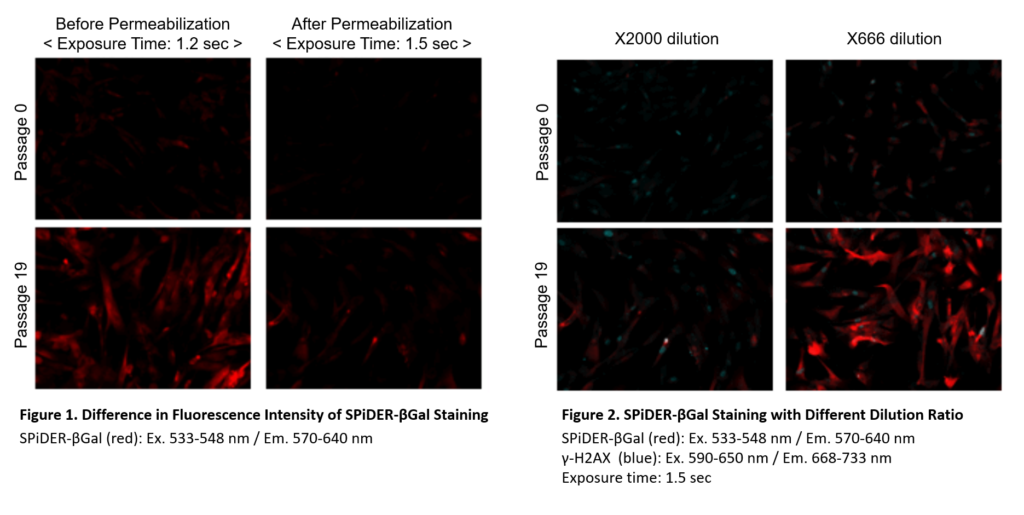
Preparation of SPiDER-βGal working solution
Dilute the SPiDER-βGal DMSO stock solution 2,000 times *1 with McIlvaine buffer (pH 6.0).
*1 Fixation and permeablization could leads to lower sensitivity (Figure 1), if you need higher signals,dilute the SPiDER-βGal DMSO stock solution 500 – 1,000 times with the McIlvaine buffer (Figure 2).
Preparation of McIlvaine buffer (pH 6.0)
Mix 0.1 mol/l citric acid solution (3.7 ml) and 0.2 mol/l sodium phosphate solution (6.3 ml). Confirm the pH is 6.0. If the pH is not 6.0, adjust the pH by adding either citric acid solution or sodium phosphate solution. Dilute this buffer 5 times with ultrapure water.
Staining procedure (35 mm dish)
1. Prepare cells on 35 mm dish for assay and culture the dish at 37℃ overnight in a 5% CO2 incubator.
2. Remove the culture medium. Add 2 ml of 4% paraformaldehyde (PFA) /PBS solution to the cells and incubate at room temperature for 3 minutes *2.
*2 Avoid a longer treatment period, which leads to decrease in SA-β-gal activity.
3. Remove the supernatant, and wash the cells 3 times with 2 ml of PBS.
4. Add 2 ml of SPiDER-βGal working solution and incubate at 37℃ for 30 minutes*3.
*3 We recommend not to use a 5% CO2 incubator for fixed cell experiments. If incubation is done in a 5% CO2 incubator, the pH of the buffer may become acidic. Acidic pH results in higher background from the endogenous β-galactosidase activity and it would be difficult to distinguish between normal cells and senescent cells.
5. After removing the supernatant, wash the cells twice with PBS.
6. Add 0.1% Triton X-100/PBS to cells and incubate for 30 minutes at room temperature.
7. Wash the cells twice with PBS.
8. Add 1% BSA/PBS to the cells and incubate for 1 hour at the room temperature
9. Add anti- γ-H2AX antibody (mouse) diluted with 1% BSA/PBS to the cells and incubate at 4℃ overnight.
10. Wash the cells 3 times with PBS.
11. Add anti- mouse secondary antibody (Cy5) diluted with 1% BSA/PBS to the cells and incubate at room temperature for 1 hour.
12. Wash cells twice with PBS and observe under a fluorescence microscope.
References
| No. | Sample Type | Instrument | Citation(Link) |
|---|---|---|---|
| 1) | Cell(HEK) Gene(LacZ) |
Microscopy Flow Cytometry |
T. Doura, M. Kamiya, F. Obata, Y. Yamaguchi, T. Y. Hiyama, T. Matsuda, A. Fukamizu, M. Noda, M. Miura, Y. Urano, "Detection of LacZ-Positive Cells in Living Tissue with Single-Cell Resolution.", Angew Chem Int Ed Engl., 2016, 55, 33 |
| 2) | Tissue (Mouse Adipose Tissue) |
Microscopy | T. Sugizaki, S. Zhu, G. Guo, A. Matsumoto, J. Zhao, M. Endo, H. Horiguchi, J. Morinaga, Z. Tian, T. Kadomatsu, K. Miyata, H. Itoh & Y. Oike, "Treatment of diabetic mice with the SGLT2 inhibitor TA-1887 antagonizes diabetic cachexia and decreases mortality", Nature Partner Journal:Aging and Mechanisms of Disease., 2017, doi:10.1038/s41514-017-0012-0. |
| 3) | Cell (HSP27-knockdown) Protein |
Microscopy | A. Park, I. Tsunoda and O. Yoshie, "Heat shock protein 27 promotes cell cycle progression by down-regulating E2F transcription factor 4 and retinoblastoma family protein p130", J. Biol. Chem., 2018, doi: 10.1074/jbc.RA118.003310 . |
| 4) | Cell (A549) | Microscopy Flow Cytometry |
R. Tanino, Y. Tsubata, N. Harashima, M. Harada and T. Isobe, "Novel drug-resistance mechanisms of pemetrexed-treated non-small cell lung cancer", Oncotarget., 2018, 9, (24), 16807. |
| 5) | Cell(NHDF) | Microscopy | Y. Kitahiro, A. Koike, A. Sonoki, M. Muto, K. Ozaki and M. Shibano. , "Anti-inflammatory activities of Ophiopogonis Radix on hydrogen peroxide-induced cellular senescence of normal human dermal fibroblasts.", J Nat Med., 2018, 72, 905. |
| 6) | Tissue (Aged Mouse Intestinal Epithelium Organoid) |
Microscopy | R. Uchida, Y. Saito, K. Nogami, Y. Kajiyama, Y. Suzuki, Y. Kawase, T. Nakaoka, T. Muramatsu, M. Kimura and H. Saito, "Epigenetic silencing of Lgr5 induces senescence of intestinal epithelial organoids during the process of aging", NPJ Aging Mech Dis., 2018, doi:10.1038/s41514-018-0031-5. |
| 7) | Tissue (Frozen Kidney Slide) |
Microscopy | S. R. Kim, A. Eirin, X. Zhang, A. Lerman and L. O. Lerman, "Mitochondrial Protection Partly Mitigates Kidney Cellular Senescence in Swine Atherosclerotic Renal Artery Stenosis.", Cell. Physiol. Biochem., 2019, 52, 617. |
| 8) | Cell(HN6, HN12, HN13) | Flow Cytometry | Liana P. Webber, Veronica Q. Yujra, Pablo A. Vargas, Manoela D. Martins. Cristiane H. Squarize, Rogerio M. Castilho, "Interference with the bromodomain epigenome readers drives p21 expression and tumor senescence", Cancer Letters., 2019, doi.org/10.1016/j.canlet.2019.06.019. |
| 9) | Cell(UE7T-13) | Flow Cytometry | H. Ise, K. Matsunaga, M. Shinohara and Y. Sakai, Improved Isolation of Mesenchymal Stem Cells Based on Interactions between N-Acetylglucosamine-Bearing Polymers and Cell-Surface Vimentin ", Stem Cells Int., 2019, 4341286, 13. |
| 10) | Cell(Mouse Corneal stroma) | Flow Cytometry | X. Wang, M. Qu, J. Li, P. Danielson, L. Yang and Q. Zhou, Induction of Fibroblast Senescence During Mouse Corneal Wound Healing.", Invest. Ophthalmol. Vis. Sci., 2019, 60, (10), 3669. |
| 11) | Cell(VZ/SVZ) | Flow Cytometry | Y. Nakatani, H. Kiyonari and T. Kondo, Ecrg4 deficiency extends the replicative capacity of neural stem cells in a Foxg1-dependent manner.", Development., 2019, 146, (4), 18. |
| 12) | Cell(HaCaT, HEK001) | Microscopy | Y. S. Ryu, K. A. Kang, M. J. Piao, M. J. Ahn, J. M. Yi, G. Bossis, Y. M. Hyun, C. O. Park and J. W. Hyun, Particulate matter-induced senescence of skin keratinocytes involves oxidative stress-dependent epigenetic modifications", Exp. Mol. Med., 2019, 51, 108. |
| 13) | Cell(HT1080) | Microscopy | E. M. Angela Ibler, E. Mohamed, L. N. Kathryn, A. B. Natalia, F. E. K. Sherif and H. Daniel, Typhoid toxin exhausts the RPA response to DNA replication stress driving senescence and Salmonella infection', Nat Commun., 2019, 10, 4040. |
| 14) | - (Review) | Flow Cytometry | B.L. Torres, A. Estepa-Fernandez, M. Rovira, M. Oraez, M. Serrano, R. Martinez-Manez and F. Sancenon,"The chemistry of senescence.", The chemistry of senescence., 2019, (3), 426-411 |
| 15) | Cell(PC12) | Microscopy | N. Wang, H. Wang, L. Li, Y. Li and R. Zhang, "β-Asarone Inhibits Amyloid-β by Promoting Autophagy in a Cell Model of Alzheimer's Disease.", Front Pharmacol., 2020, 10, 1529 |
| 16) | Cell(A2780) | Flow Cytometry | Z. Wang, J. Gao, Y. Ohno, H. Liu and C. Xu, "Rosiglitazone ameliorates senescence and promotes apoptosis in ovarian cancer induced by olaparib.", Cancer Chemother Pharmacol., 2020. |
| 17) | Tissue (Mouse Frozen Kidney Slide) |
Microscopy Flow Cytometry |
J. H. Cho, E. Kim, Y. Son, D. Lee, Y. S. Park, J. H. Choi, K. Cho, K. Kwon and J. Kim, "CD9 Induces Cellular Senescence and Aggravates Atherosclerotic Plaque Formation.", Cell Death Differ., 2020, doi: 10.1038/s41418-020-0537-9 |
| 18) | Cell(T cell) | Flow Cytometry | S. Yoshida, H. Nakagami, H. Hayashi, Y. Ikeda, J. Sun, A. Tenma, H. Tomioka, T. Kaawano, M. Shimamura, R. Morishita and H. Rakugi, "The CD153 vaccine is a senotherapeutic option for preventing the accumulation of senescent T cells in mice.", Nat. Commun., 2020, 11, (2482), doi:10.1038/s41467-020-16347-w |
| 19) | Cell(PC12) | Microscopy | N. Wang, H. Wang, L. Li, Y. Li and R. Zhang, "β-Asarone Inhibits Amyloid-β by Promoting Autophagy in a Cell Model of Alzheimer's Disease.", Front Pharmacol., 2020, 10, 1529 |
| 20) | Cell(ARPE-19) | Microscopy | T. Yamazaki, H. Suzuki, S. Yamada, K. Ohshio, M. Sugamata, T. Yamada and Y. Morita, "Lactobacillus paracasei KW3110 Suppresses Inflammatory Stress-Induced Premature Cellular Senescence of Human Retinal Pigment Epithelium Cells and Reduces Ocular Disorders in Healthy Humans”, Int J Mol Sci, 2020, 21(14), 5091 |
| 21) | Cell (Epithelial cells of zebrafish) |
Microscopy | Y. Haraoka, Y. Akieda, Y. Nagai, C. Mogi and T. Ishitani, "Zebrafish imaging reveals TP53 mutation switching oncogene-induced senescence from suppressor to driver in primary tumorigenesis", Nat. Commun., 2022, doi:10.1038/s41467-022-29061-6. |
| 22) | Tissue (Adipose) | Microscopy | A. Kita, Y. Saito, N. Miura, M. Miyajima, S. Yamamoto, T. Sato, T. Yotsuyanagi, M. Fujimiya and T. Chikenji, "Altered regulation of mesenchymal cell senescence in adipose tissue promotes pathological changes associated with diabetic wound healing", Commun. Biol., 2022, doi:10.1038/s42003-022-03266-3. |
| 23) | Cell (hMPC) | Microscopy | X. Liu, Z. Liu, Z. Wu, J. Ren, Y. Fan, L. Sun, G. Cao, Y. Niu, B. Zhang, Q. Ji, X Jiang, C. Wang, Q. Wang, Z. Ji, L. Li, C. R. Esteban, K. Yan, W. Li, Y. Cai, S. Wang, A. Zheng, Y. E. Zhang, S. Tan, Y. Cai, M. Song, F. Lu, F. Tang, W. Ji, Q. Zhou, J. Belmonte, W. Zhang, J. Qu, G. Liu, "Resurrection of endogenous retroviruses during aging reinforces senescence", Cell, 2023, doi:10.1016/j.cell.2022.12.017. |
Q & A
-
Q
What is the approximate number of uses per kit?
-
A
Please refer to the following for the number of samples that can be measured.
35 mm dish Micro plate Chamber slide Dish type 
Number of didhes 10 dishes 2 plates 7 slides Note: The above number will vary depending on the amount of staining solution added to each well. Please check the dish used and the required amount of staining solution before use.
-
Q
What is the reason of adding Bafilomycin A1 (Baf. A1)?
-
A
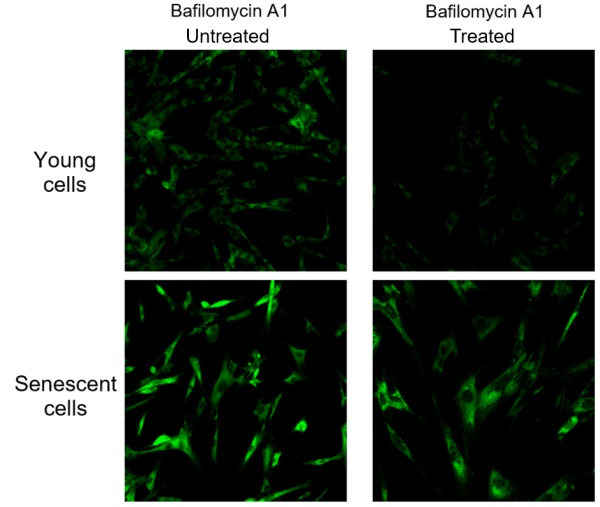
Endogenous β-galactosidase existing in living cells interfere with selective detection of SA-β-Gal. Baf. A1 is an inhibitor of ATPase in lysosome. pH in lysosome is kept neutral by adding Baf. A1. Cellular Senescence Detection Kit - SPiDER-βGal contains Baf. A1 which allows to detect SA-β-Gal selectively. Baf. A1 is utilized for living cell assays only. Baf. A1 is not used in fixed cells because intracellular pH is controlled with the buffer.
The figure above shows the difference in SA-β-Gal detection with and without Baf. A1. Baf. A1 is also known as an autophagy inhibitor. When using Baf. A1. on live cells, please consider whether there are any effects on the experiment and if so, we recommend experiments with fixed cells. Baf. A1 is not used with fixed cells as the intracellular pH is controlled by buffer. Fixed cells are stained with SPiDER-βGal as procedure described in the manual.
-
Q
How stable is DMSO stock solution?
-
A
SPiDER-βGal DMSO stock solution and Bafilomycin A1 DMSO stock solution is stable for 1 month at -20 ℃.
-
Q
How stable is working solution?
-
A
SPiDER-βGal working solution and Bafilomycin A1 working solution can’t be stored. Be sure to use the working solution immediately.
-
Q
Are there any advices when observing the senescent cells?
-
A
Lipofuscin is a fluorescent pigment that accumulates in a variety of cell types with age. Lipofuscin consists of autofluorescent granules and may results in high background for fluorescence microscopy. In order to achieve accurate SA-β-gal activity assay in senescent cells, we recommend to prepare samples without SPiDER-βGal staining as a negative control. Please compare fluorescence intensity of both cells with or without SPiDER-βGal staining.
> For Flow Cytometry Detection
Step 1. Prepare senescent cells and non-senescent cells. Measure MFI (Mean Fluorescence Intensity) of samples below.
[Senescent cells]
Sample A: The cells stained with SPiDER-βGal
Sample B: The cells without SPiDER-βGal staining
[Non-senescent cells]
Sample A’: The cells stained with SPiDER-βGal
Sample B’: The cells without SPiDER-βGal staining
Step 2. Calculate SA-β-gal activity (senescent cells) with the following formula
SA-β-gal activity (senescent cells) = MFI of Sample A - MFI of Sample B
Step 3. Calculate SA-β-gal activity (non-senescent cells) with the following formula
SA-β-gal activity (non-senescent cells) = MFI of Sample A’ - MFI of Sample B’
- Determine the SA-β-gal activity by comparing the SA-β-gal activity between senescent cells and non-senescent cells.
- Change of SA-β-gal activity associated with senescence = (Value from Step 2- value from Step 3)
>For Microscopy
Step 1. Prepare senescent cells without SPiDER-βGal staining and observe fluorescent image.
Step 2. Adjust detection sensitivity in microscopy to reduce background autofluorescence of lipofuscin.
Step 3. Observe fluorescent image of senescent cells and non-senescent cells under the settled condition in step 2.
-
Q
Can SPiDER-βGal be used with fixed cells?
-
A
It is possible. If fixed cells are stained with SPiDER-βGal, it is not necessary to pre-treat the cells with bafilomycin A1. However, McIlvain buffer adjusted to pH 6.0 must be prepared separately. See manual for details.
-
Q
What is the protocol for detecting fixede and stained cells using a flow cytometer?
-
A
See protocol below.
(1) Prepare cells in 35 mm dishes and incubate overnight at 37°C in a 5% CO2 incubator.
(2) After removing the culture medium, wash the cells once with 2 ml of HBSS.
(3) Treat the cells with trypsin and mix with 500 µl serum containing medium to collect the cells.
(4) Centrifuge at 300 xg for 5 minutes and remove the supernatant.
(5) Suspend the cells in 100 µl 2% PFA/PBS to the cells and incubate for 5 minutes at room temperature.
(6) Centrifuge at 300 xg for 5 minutes and remove the supernatant.
(7) Suspend the cells in 500 µl HBSS, centrifuge at 300 ×g for 5 minutes and remove the supernatant. Repeat this step twice.
(8) Add 500 µL of SPiDER-βGal working solution (for fixed cells) and incubate for 30 minutes at 37°C.
Note: Do not use a 5% CO2 incubator due to reduce pH fluctuations.
(9) Centrifuge at 300 xg for 5 minutes and remove the supernatant.
(10) Suspend the cells in 500 µl HBSS, centrifuge at 300 ×g for 5 minutes and remove the supernatant. Repeat this step twice.
(11) Suspend the cells in 500 µl HBSS and analyze on a flow cytometer.
-
Q
Can SPiDER-βGal staining and immunostaining be performed after fixation of the cells?
-
A

Preparation of SPiDER-βGal working solution
Dilute the SPiDER-βGal DMSO stock solution 2,000 times *1 with McIlvaine buffer (pH 6.0).
*1 Fixation and permeablization could leads to lower sensitivity (Figure 1), if you need higher signals,dilute the SPiDER-βGal DMSO stock solution 500 – 1,000 times with the McIlvaine buffer (Figure 2).Preparation of McIlvaine buffer (pH 6.0)
Mix 0.1 mol/l citric acid solution (3.7 ml) and 0.2 mol/l sodium phosphate solution (6.3 ml). Confirm the pH is 6.0. If the pH is not 6.0, adjust the pH by adding either citric acid solution or sodium phosphate solution. Dilute this buffer 5 times with ultrapure water.Staining procedure (35 mm dish)
1. Prepare cells on 35 mm dish for assay and culture the dish at 37℃ overnight in a 5% CO2 incubator.
2. Remove the culture medium. Add 2 ml of 4% paraformaldehyde (PFA) /PBS solution to the cells and incubate at room temperature for 3 minutes *2.
*2 Avoid a longer treatment period, which leads to decrease in SA-β-gal activity.
3. Remove the supernatant, and wash the cells 3 times with 2 ml of PBS.
4. Add 2 ml of SPiDER-βGal working solution and incubate at 37℃ for 30 minutes*3.
*3 We recommend not to use a 5% CO2 incubator for fixed cell experiments. If incubation is done in a 5% CO2 incubator, the pH of the buffer may become acidic. Acidic pH results in higher background from the endogenous β-galactosidase activity and it would be difficult to distinguish between normal cells and senescent cells.
5. After removing the supernatant, wash the cells twice with PBS.
6. Add 0.1% Triton X-100/PBS to cells and incubate for 30 minutes at room temperature.
7. Wash the cells twice with PBS.
8. Add 1% BSA/PBS to the cells and incubate for 1 hour at the room temperature
9. Add anti- γ-H2AX antibody (mouse) diluted with 1% BSA/PBS to the cells and incubate at 4℃ overnight.
10. Wash the cells 3 times with PBS.
11. Add anti- mouse secondary antibody (Cy5) diluted with 1% BSA/PBS to the cells and incubate at room temperature for 1 hour.
12. Wash cells twice with PBS and observe under a fluorescence microscope.
-
Q
Is there any advice if the fluorescence of stained cells is low?
-
A
Please check the following points.
(1) Use the filter that matches the fluorescence characteristics of the reagent.
<Recommended filters>
Fluorescence microscope: excitation (500-540 nm), fluorescence (530-570 nm)
Flow cytometer: excitation (488 nm), fluorescence (500-540 nm)
(2) Use fresh working solution(3) Increase the staining time.
If fluorescence cannot be confirmed after 30 minutes of incubation after addition of SPiDER-βGal working solution, increase the incubation time to 45-60 minutes.
-
Q
Can the cells be fixed after staining with SPiDER-βGal?
-
A
It is possible. Fixation with 4% paraformaldehyde is recommended.
-
Q
Does serum or phenol red in the medium affect detection?
-
A
Serum and phenol red in the medium do not affect the detection of SA-β-gal.
-
Q
If there is no difference in fluorescence intensity between senescent and control cells, what should be checked?
-
A
-
Q
No difference in fluorescence intensity between senescent cells and control cells
-
A
Cellular senescence wasn’t induced. Please prepare for positive control. Please visit product page and check “Positive Control”
-
Q
Background in fix cells assay
-
A
The incubation with SPiDER-βGal working solution was done in a 5% CO2 incubator. We recommend not to use a 5% CO2 incubator during incubation with SPiDER-βGal working solution. If incubation is done in a 5% CO2 incubator, the pH of the buffer may become acidic. Acidic pH results in higher background from the endogenous β-galactosidase activity and it would be difficult to distinguish between control cells and senescent cells. Please incubate the plate in a dry incubator without CO2 .
Handling and storage condition
| 0-5°C |